Abstract
Several Aureobasidium spp. strains isolated from wild environments during winter 2022 were characterized by sequence analysis of the internal transcribed spacer (ITS), the translation elongation factor EF-1α gene (EF1), and part of the elongase gene (ELO). The variability in the EF1 and ELO loci are higher than in the ITS. All strains but one (UC14), were identified as A. pullulans. To assess the effectiveness of the characterized strains as biocontrol agents (BCAs) of diseases occurring during postharvest storage, a selection of the strains was evaluated by in vitro and in vivo assays. On average, the reduction of Monilinia spp. colony growth was more marked for non-volatile metabolites than for volatile (VOCs). Strain UC14 provided the strongest mycelial growth reduction of Monilinia fructicola by VOCs (66%). According to the in vivo results, all strains were effective in controlling brown rot during cold storage and remarkably in restricting the growth of Monilinia polystroma. In particular, VB23 was the most effective in controlling brown rot incidence, by 80%, 60%, 100%, and severity, by 79.5%, 72.7% and 100%, for Monilinia laxa, M. fructicola, and M. polystroma, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The increasing limitation to the use of pesticides, the allowable limit of residues on fruits, and the risk of development of resistant strains together with the request of the market for a more sustainable fruit production, is leading plant defence science into a change (Casals et al., 2022). The new European plant health regulation (EU 2016/2031) jointly with the “European Green Deal” aims to accelerate the transition to a more sustainable food system recommending a reduction by 50% of pesticides and an increment of the land used for organic by 25% farming by 2030. A significant number of pesticides are presently used to prevent brown rot on stone fruits, caused by Monilinia laxa (Aderh. & Ruhland) Honey, Monilinia fructicola (Winter) Honey, and Monilinia polystroma Kohn. These species are notorious pathogens of rosaceous plants all around the world (Petróczy et al., 2012). Fruits that has been infected in the field can lead to losses during the postharvest phase in packing houses (Larena et al., 2005a). In Europe, M. laxa is the prevalent species (Larena et al., 2005b) but since the detection of M. fructicola in France (EPPO, 2002), Spain (de Cal et al., 2009), Switzerland (Bosshard et al., 2006), Hungary (Petróczy & Palkovics, 2009), Italy (Pellegrino et al., 2009) and M. polystroma (Côté & Meldrum, 2004;Petróczy & Palkovics, 2009; EPPO, 2011; Martini et al., 2014) on peaches, it is now apparent that these species can co-exist (Villarino et al., 2013).
A sustainable solution to limit brown rot diseases during storage is biological control by exploiting microorganisms (Droby et al., 2009) referred to as biocontrol agents (BCAs). Recent studies proved the efficacy of Aureobasidium pullulans (de Bary) G. Arnaud in controlling some of the most detrimental pathogens of fruits in the postharvest phase (Di Francesco et al., 2018, 2020a, 2020b; Ippolito et al., 2000; Mari et al., 2012,). Aureobasidium pullulans is a polyextremotolerant black yeast, epiphyte or endophyte, globally distributed, found in tropical, temperate, and polar areas (Di Francesco et al., 2023; Gostinčar et al., 2014). Other interesting species within the Aureobasidium genus are A. melanogenum, A. subglaciale, and A. namibiae (Gostinčar et al., 2014), although A. melanogenum could be associated to human infections, being able to grow at 37 °C (Di Francesco et al., 2020b).
Unfortunately, not much data is available regarding the biocontrol ability of A. namibiae, a species isolated from extreme environments and able to tolerate up to 10% of NaCl and to grow between 10 °C and 30 °C (Zalar et al., 2008; Gostincar et al., 2014). Such features make it a promising source for new antimicrobial metabolites and a candidate for applications in agriculture. Recently, new species such as Aureobasidium vineae, Aureobasidium mustum, Aureobasidium uvarum (Onetto et al., 2020), Aureobasidium castaneae (Jiang et al., 2021), Aureobasidium aerum (Cheng et al., 2022) and Aureobadidium acericola (Lee et al., 2021) were described on the ground of phylogenetic and morphological analyses, but their potential as BCAs has not been assessed yet (Di Francesco et al., 2023).In general, the most studied environment for isolation of antagonistic yeasts to be applied during the postharvest phase, is the fruit surface (Mari et al., 2012; Parafati et al., 2015). In general, the isolation of wild yeasts from fruit surface holds a great promise for disease control in postharvest, although the biocontrol ability of the isolated strains may vary greatly, and hence needs careful assessment.
In order to improve the knowledge on the potential of wild yeasts, we undertook the present work with the aim to molecularly characterize the Aureobasidium spp. population from wild environments sampled during the cold season and evaluate the biocontrol ability of selected strains through in vitro (co-culturing, non-volatile and volatile metabolites assays) and in vivo assays on apricots against Monilinia spp.
Material and methods
Microorganisms’ isolation and morphological identification
Thirty-seven wild black yeasts were isolated during winter 2022 from wild plants located in Friuli Venezia Giulia, Italy. Olive, laurel, plum, and viburnum healthy leaves were sampled. Plants material (2 g) was washed with 20 mL of sterile distilled water (SDW) amended with Tween 20 (0.05% v/v, Sigma-Aldrich, USA), collected in sterile flasks subsequently placed at 20 °C for 1 h under continuous stirring on a rotary shaker (250 rpm). Samples were centrifuged for 30 min at 3500 rpm and the supernatant was discarded. The pellet was resuspended with 1 mL of SDW and then 100 μL of each solution wase spread on Petri dishes containing NYDA medium (non-selective general-purpose medium for the cultivation of microorganisms) (8 g L−1 Nutrient broth, 5 g L−1 Yeast extract, 10 g L−1 Dextrose, and 25 g L−1 of technical Agar) (Oxoid, UK). Plates were incubated for 48 h at 20 °C. Only single colonies displaying A. pullulans morphological characteristics like smooth, slimy, pale pink or cream color, as reported by Barnett and Barry (1998), were individually isolated on new plates and then purified. Pure cultures were observed with a microscope (Zeiss AXIO Observer.Z1). Only the strains with common Aureobasidium morphological characteristics such as hyaline, thin-walled and septate hyphae, and ellipsoidal, variable in shape and dimension, hyaline conidia were retained, according to Zalar et al. (2008).
Fifteen Monilinia spp. strains were isolated from symptomatic apricots collected in the experimental orchards of the University of Udine. Purified fungal isolates were grown on PDA (Potato Dextrose Agar, Oxoid, UK) plates for 5 d at 20 °C. Yeast isolates were processed for DNA extraction and molecular identification. For the subsequent experiments, yeast cells and fungal conidia were collected and suspended in SDW containing 0.05% (v/v) Tween 20 and the suspensions were adjusted to 1 × 108 cells mL−1 and 1 × 105 conidia mL−1, respectively, by using a hemocytometer. Before proceeding to the in vivo assay, fungal strain suspensions (1 × 105 conidia mL−1) were wound-inoculated on apricots to test their pathogenicity.
Fruits
Apricots (Prunus armeniaca L.) cv “Bella d’Imola” (12°Brix) were harvested in an experimental orchard of the University of Udine (Italy) (46°01′56.4"N 13°13′23.6"E) and immediately processed. Fruits homogeneous in size and injuries-free were selected.
Molecular characterization of Aureobasidium spp. and Monilinia spp
Yeast isolates were grown on flasks containing NYDB (NYDA without agar). Cellular cultures were centrifuged at 3500 rpm for 30 min and the supernatant was discarded. Cells were frozen with liquid nitrogen and then stored at – 80 °C. The genomic DNA extraction was conducted by using 150 mg of each sample collected in 2 mL microcentrifuge tubes (Eppendorf AG, Hamburg, Germany) and by following the protocol described by Carlucci et al. (2013).
For molecular characterization of Aureobasidium spp., a multi-locus approach was used. The nucleotide sequences of Internal Transcribed Spacer (ITS), translation elongation factor EF-1α gene (EF1) and part of the elongase gene (ELO) were determined as described by Zalar et al. (2008). The ITS region was PCR amplified by using the primers ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTATTGATATGC-3’) described by White et al. (1990). Primers ELO2-F(5’-CACTCTTGACCGTCCCTTCGG-3’) and ELO2-R (5’-GCGGTGATGTACTTCTTCCACCAG-3’) (Zalar et al., 2008) were used for the PCR amplification of the elongase gene. Finally, for the amplification of the elongation factor 1α gene, EF1-728F (5’-CATCGAGAAGTTCGAAGG-3’) and EF1-986R (5’-TACTTGAAGGAACCTTTACC-3’) (Carbone & Kohn, 1999) were used. PCR amplifications were carried out separately on a MiniAmp Plus thermal cycler (Thermo Fisher Scientific, USA). For the ITS region, an initial denaturation of 94 °C for 2 min was followed by 40 cycles of 94 °C for 40 s, 55 °C for 40 s, and 72 °C for 1 min, with a final extension of 5 min at 72 °C. For ELO and EF1, the parameters were slightly different: an initial denaturation of 94 °C for 5 min, 35 cycles of 94 °C for 15 s, 56 °C for 40 s and 40 s at 72 °C, with a final extension of 8 min at 72 °C. The PCR products were purified and sequenced by BMR Genomics (Padova, Italy).
For Monilinia spp. isolates identification, DNA was extracted as above,after collecting 150 mg of mycelium scraped off the PDA plates after 7 days of growth. DNA was amplified by using primers UniMon_Rev (5’-GAGCAAGGTGTCAAAACTTCCAT-3’) and UniMon_Forw (5’-ATCGGCTTGGGAGCGG-3’) (Petróczy et al., 2012), based on the different lengths of the ITS. The amplified region is 397 bp for M. laxa, 417 bp for M. fructigena, 433 bp for M. polystroma, and 594 bp for M. fructicola (Petróczy et al., 2012).
RAPD analysis
To corroborate the findings concerning the strain diversity highlighted among the isolates by gene sequencing, DNA of 8 strains, selected as the most representative of each strain cluster, was subjected to Random Amplification of Polymorphic DNA (RAPD) analysis. The protocol described by Di Francesco et al. (2018) was used, and the purified genomic DNA was subjected to PCR for RAPD analysis by using a MiniAmp Plus thermal cycler (Thermo Fisher Scientific, USA). Four random primers were used (Supplemental material S1) and PCR products were separated and visualized on 1% agarose gel.
In vitro assays
To evaluate the effectiveness of the selected strains as BCAs, three different in vitro experiments were conducted as reported by Di Francesco et al. (2017). The interaction between the yeast strains and fungal pathogens belonging to three different species (M. laxa AML1, M. fructicola AMF1, M. polystroma AMP1) was evaluated by co-culturing assays. On PDA plates (90 mm diameter), a mycelial plug of each pathogen (6 mm diameter, from 7-d old colony) was placed 25 mm from the edge of the plate, and a streak of each yeast strain (from a 48-h old culture) was placed 25 mm from the other side of the edge of the plate. The non-volatile organic compounds (non-VOCs) assays were carried out by using sterile cellophane layers (BIORAD, USA), positioned on PDA plates, on which 100 μL of each yeast suspension (1 × 108 cells mL−1) were spread. After 48 h, the cellophane layer was removed and a mycelial plug from 7 d old colony of each pathogen species (6 mm diameter) was placed in the centre of the plate (Di Francesco et al., 2020a). For the evaluation of BCAs VOCs antifungal activity, a double Petri dish assay was performed according to the method of Rouissi et al. (2013) as modified by Di Francesco et al. (2020b).
The plates were incubated for 5 days at 25 °C for M. laxa and M. fructicola and for 14 d at 20 °C in the case of M. polystroma (Di Francesco et al., 2017). The sample unit of each assay consisted in 6 plates for each interaction. The experiments were conducted twice. The inhibition rate of mycelial growth was calculated using the equation (Dennis & Webster, 1971):
where d1 and d2 are the control colony and the treated colony diameters, respectively.
In vivo assay
Apricots were disinfected by immersion for 1 min in sodium hypochlorite solution (1%) and washed twice in sterile water. Fruits were wounded (2 × 2 × 2 mm) on the equatorial area with a sterile needle and inoculated with 20 μL of each selected yeast suspension (UC14, VB23, UC11, UOR18, UOR112, SE0, AL25, UC12) (1 × 108 cells mL−1). After the complete adsorption (1 h), 20 μL of conidial suspension (1 × 105 conidia mL−1) of each of the three pathogens (AML1, AMF1, AMP1) were pipetted into the wounds. After air drying, fruits were stored at 0 °C and 90% R.H. for 30 d in plastic boxes. The percentage of rotten fruits was detected after shelf-life of 7 d at 20 °C. Fruits inoculated with SDW (20 μL) and Scholar® (active substance fludioxonil, 2 mL/L) (Syngenta, Switzerland) were controls. Each treatment was represented by 20 fruits, and there were three replicates per treatment. The experiment was conducted twice.
Data analysis
All the obtained data from in vitro and in vivo experiments were subjected to a one-way analysis of variance (ANOVA). The statistical comparisons of the means were established by using Tukey’s HSD test (α = 0.01). All the analyses were conducted by using Minitab 17® statistical software (Minitab, USA). DNA sequences were aligned by using Clustal Omega (Sievers et al., 2011) and the phylogenetic trees were built from pairwise distances using Seaview 5.0.5 (Gouy et al., 2010). The Neighbor-Net graph was built by using Split Trees Version 4 (Huson & Bryan, 2006). The same software tool was used to build and display a tree based on DNA fragment presence/absence data from RADP patterns as detected on agarose gels.
Results
Characterization and identification of the strains
From a first screening of the microorganisms grown on NYDA plates, thirty-seven colonies morphologically referable to Aureobasidium by microscope identification were selected, isolated on new agar plates, and purified. All the pure cultures were molecularly analysed.
The amplification of the target genes produced sequences of 508, 691, and 230 bp respectively for ITS, ELO, and EF-1alpha. According to the analysis, thirty-six strains were identified as A. pullulans, while one strain (UC14) was found to belong to the Aureobasidium genus but could not be unambiguously ascribed to a species, as reported in Table 1. The analysis carried out on the ITS region showed no differences among the strains (Fig. 1A), with the exception of the sequence of strain UC14 that displayed seven different nucleotides different from the others. Conversely, the variability at the ELO and EF1 loci was larger and more discriminant than that of the ITS, in particular for strain UC14 (on average, 65 and 44 nts difference for ELO and EF1 genes, respectively, vs. the remaining strains). Overall, the analysis of the sequences of the ELO gene revealed 8 distinct clusters of strains (Fig. 1B). The groups arising from the ELO gene sequence analysis did not match with those based on EF1 gene (Fig. 1C). This issue could probably be explained by the presence of a recombinant population (Fig. 1D).
One strain from each group (AL25, SE0, UC11, UC14, UC22, UOR112, UOR18, VB23), based on the ELO gene, was selected and the relative diversity was assessed by RAPD (A9) with electrophoretic separation (Fig. 2). The analysis of the RAPD patterns supported the notion of a diversified sample of strains, with strains UC14 and UC11 that differ the most from the others (Fig. 3).
Regarding the identification of Monilinia spp. isolates, the most represented species was M. laxa (13 isolates). The remaining strains were identified as either M. fructicola (1 isolate) and M. polystroma (1 isolate) (data not shown).
In vitro assays
Strains isolated in this work were evaluated through different assays. Regarding the co-culturing assay, all the selected strains provided on average a growth inhibition of AML1 and AMF1 by 23% and 25%, respectively, with no significant difference between the tested strains. Monilinia polystroma AMP1 strain was the tested fungal pathogen inhibited to the highest degree (on average 40%), in particular by strains SEO, AL25, UC22 with an inhibition of 49.1%, 46.3% and 50%, respectively, as compared to the control (Fig. 4).
Co-culturing assay between the selected Aureobasidium spp. strains and Monilinia spp. Colony diameters (mm) were measured after 5 days at 25 °C for M. laxa and M. fructicola, and after 14 days at 20 °C for M. polystroma. Each value is the mean of 6 plates (replicates) ± standard error. Different letters represent significant differences among the strains according to Tukey’s Test (α = 0.01)
In the case of non-volatile assay, strains UC14, UOR18, AL25, and UC22 completely inhibited M. polystroma AMP1 mycelial growth (Fig. 5). The mycelial growth of both M. laxa AML1 and M. fructicola AMF1 was reduced by 70%, on average, by the tested strains. Noticeably, VB23 inhibited by 76% AML1 and UC11 by 81% AMF1.
Effect of non-volatile metabolites produced by selected isolates on the mycelial growth (mm) of Monilinia spp. Colony diameter (mm) was measured after 5 days at 25 °C for M. laxa and M. fructicola, and after 14 days at 20 °C for M. polystroma. Each value is the mean of 6 plates (replicates) ± standard error. Different letters indicate significant differences according to Tukey’s Test (α = 0.01)
The efficacy of VOCs produced by the tested strains was strictly dependent on the targeted pathogen species. As shown in Fig. 6, AML1 compared to AMF1, while AMP was less affected by the VOCs produced by the wild yeast strains. In fact, on average, the percentage of reduction detected for AML1, AMF1, and AMP1 was 28.6%, 32.8%, and 40.3%, respectively. Globally, isolates UC14 and AL25 resulted the most effective strains according to bio-active VOCs production results.
Effect of volatile compounds produced by selected Aureobasidium spp. strains on the mycelial growth (mm) of Monilinia spp. Colony diameter (mm) was measured after 5 days at 25 °C for M. laxa and M. fructicola, and after 14 days at 20 °C for M. polystroma. Each value is the mean of 6 plates (replicates) ± standard error. Different letters indicate significant differences according to Tukey’s Test (α = 0.01)
In vivo assay
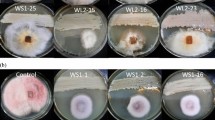
A relevant inhibitory activity of the tested strains was detected by in vivo assays (Fig. 7). It was evidenced that isolate VB23 was the most effective strain in controlling brown rot (Supplemental material S2) reducing by 80%, 60%, 100% the incidence (data not reported) and by 79.5%, 72.7% and 100% the severity of brown rot caused by AML1, AMF1, AMP1, respectively, as compared to the control.
Antagonistic effect by Aureobasidium spp. strains against Monilinia spp. in apricot. Fruits were artificially inoculated with antagonist cells and conidial suspensions (20 μL) (1 × 108 cells mL−1 and 1 × 105 conidia mL−1, respectively). Apricots were incubated at 0 °C and 90% RH for 30 days. Severity of rotten fruits was detected after 7 d at 20 °C. Fruit inoculated with water and Scholar® were used as negative and positive control treatments, respectively. According to Tukey’s HSD Test, different letters represent significant differences within the treatments among the fungal pathogens (α = 0.01)
Strain UC14 showed the same efficacy as strain VB23 in reducing AML1 severity symptoms. Strains AML1 (average lesion size: 44 mm) and AMF1 (55 mm) resulted more aggressive than AMP1 (19 mm). The synthetic product (Scholar®) was confirmed as the most effective treatment, although the effectiveness of strain VB23 was comparable to that of the chemical treatment against AMP1 strain.
Discussion
It is well known that the phenotypic plasticity Aureobasidium spp. promotes its tolerance and adaptability to different environmental conditions (Pinto et al., 2018; Di Francesco et al., 2023). The best-studied species of Aureobasidium, namely A. pullulans, has been predominantly isolated from the fruit carposphere (Bozoudi & Tsaltas, 2018). Indeed, microorganisms naturally present on the surface of fruits are the most common source of BCAs being used against postharvest fungal fruit diseases (Lima et al., 1997; Janisiewicz & Korsten, 2002; Talibi et al., 2014). In fact, strains belonging to extreme-tolerant species are potential candidates for use during cold storage.
The present study reports the identification of strain UC14, an isolate molecularly distinct from the others and likely related to the species A. namibiae, according to the results of the analysis of ITS and ELO gene sequences. Unfortunately, no EF1 gene sequences were available for A. namibie. Therefore, further investigations are needed to better characterize this strain.
Although the isolation was carried out during the winter period, no strain with similarity to A. subglaciale was isolated. With the exception of isolate UC14, there was no evidence that the population sampled in this work may significantly differ from those sampled during spring and on fruit surfaces in more conventional environments.
Cold can induce biochemical, biophysical, and physiological changes to cells (Garcia-Rios et al., 2016). Most microorganisms resist to a variety of changing conditions in their environment in order to survive. Nevertheless, the well-equipped for cold temperature growth, psychrophilic and psychrotrophic microorganisms have to make numerous adjustments when facing temperatures lower than optimum (Berry & Foegeding, 1997). Moreover, the cold shock response has not been fully delineated but seems to be an adaptive trait of A. pullulans, characterized by a high physiological adaptability.
The lack of population structure (Fig. 1A, B, C), despite the large observed diversity within all the yeast isolates, might be explained by a high recombination among A. pullulans strains (Gostincar et al., 2019).
In this study, the mechanism of action of the antibiosis was deeply investigated by in vitro assays in view of future exploitation for the control of Monilinia spp. Hence, the results obtained provided preliminary information on the effectiveness of the strains and on their most active modes of action. The results showed a higher reduction of the growth of the pathogens by non-volatile metabolites with respect to volatile metabolites, particularly in the case of M. polystroma (AMP1). These results are in agreement with other studies reporting limited efficacy of VOCs in the control of Monilinia spp. mycelial growth (Di Francesco et al., 2020a), rather than conidial germination.
Non-volatile organic compounds produced by BCAs have also been reported to inhibit other postharvest pathogens such as Alternaria alternata (Pérez-Corral et al., 2020; Wang et al., 2022). According to the literature, Aureobasidium spp. secondary metabolites involved in the antagonism against pathogens comprehend extracellular hydrolytic enzymes, such as chitinase and beta-1,3-glucanase, and various antimicrobial compounds such as aureobasidin A, exophilin A, and siderophores (Castoria et al., 2001; Zhang et al., 2010; Gostinčar et al., 2014a).
Strains UC14 and AL25 showed the best inhibition results by producing VOCs against M. fructicola mycelial growth compared to the rest of the tested yeast population. However, compared to the synthetic fungicide (Scholar®), which completely controlled the insurgency of the disease caused by all three pathogens, the BCAs exhibited lower effectiveness, and only AMP was totally controlled by the BCA, strain VB23. Other studies reported similar results with BCA effectiveness significantly lower than the synthetic fungicides (Lahlali et al., 2020; Palmieri et al., 2022). Since Aureobasidium spp. are considered highly tolerant microorganisms, a desired trait in the biocontrol application (Zajc et al., 2019), we hypothesize that the efficacy of yeasts as BCAs will be greater when applied in a stressful situation, such as the low temperature condition during fruit storage.
Based on these preliminary results, future studies will be carried out to test the effectiveness of the target yeasts in extreme storage conditions (e.g., temperature, relative humidity—R.H.) and further investigate on the possible physiological differences that exist between species isolated during cold or warm seasons.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Barnett, H. L., & Barry, B. H. (1998). Illustrated Genera of imperfect fungi (p. 0890541922). The American Phytopathological Society Press.
Berry, E. D., & Foegeding, P. M. (1997). Cold temperature adaptation and growth of microorganisms. Journal of Food Protection, 60, 1583–1594. https://doi.org/10.4315/0362-028X-60.12.1583
Bosshard, E., Hilber-Bodmer, M., Schärer, H. J., Bünter, M., & Duffy, B. (2006). First report of the quarantine brown rot pathogen Monilinia fructicola on imported stone fruits in Switzerland. Plant Disease, 90, 1554. https://doi.org/10.1094/PD-90-1554C
Bozoudi, D., & Tsaltas, D. (2018). The multiple and versatile roles of Aureobasidium pullulans in the vitivinicultural sector. Fermentation, 4, 85. https://doi.org/10.3390/fermentation4040085
Bryant, D., & Moulton, V. (2004). Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution, 21, 255–265. https://doi.org/10.1093/molbev/msh018
Carbone, I., & Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia, 91, 553–556. https://doi.org/10.1080/00275514.1999.12061051
Carlucci, A., Raimondo, M., Cibelli, F., & Phillips, A. J. (2013). Pleurostomophora richardsiae, Neofusicoccum parvum and Phaeoacremonium aleophilum associated with a decline of olives in southern Italy. Phytopathologia Mediterranea, 52, 517–527. https://www.jstor.org/stable/42685430
Casals, C., Torres, R., Teixidó, N., de Cal, A., Segarra, J., & Usall, J. (2022). Brown rot on stone fruit: From epidemiology studies to the development of effective control strategies. Scientia Horticulturae, 301. https://doi.org/10.1016/j.scienta.2022.111096
Castoria, R., De Curtis, F., Lima, G., Caputo, L., Pacifico, S., & De Cicco, V. (2001). Aureobasidium pullulans (LS-30) an antagonist of postharvest pathogens of fruits: Study on its modes of action. Postharvest Biology and Technology, 22(1), 7–17. https://doi.org/10.1016/S0925-5214(00)00186-1
Cheng, B., Wang, N., Jiang, N., Jiang, Y., Tu, Y., Yaquan, Z., Han, X., & Li, Y. (2022). Aureobasidium aerium (Saccotheciaceae, Dothideales), a new yeast-like fungus from the air in Beijing, China. Phytotaxa, 544, 185–192. https://doi.org/10.11646/phytotaxa.544.2.4
Côté, M. J., & Meldrum, A. J. (2004). Identification of Monilinia fructigena, M. fructicola, M. laxa, and Monilia polystroma on inoculated and naturally infected fruit using multiplex pcr. Plant Disease, 88, 1219–1225. https://doi.org/10.1094/pdis.2004.88.11.1219
De Cal, A., Gell, I., Usall, J., Viñas, I., & Melgarejo, P. (2009). First report of brown rot caused by Monilinia fructicola in peach orchards in Ebro Valley. Spain. Plant Disease, 93, 763. https://doi.org/10.1094/PDIS-93-7-0763A
Dennis, C., & Webster, J. (1971). Antagonistic properties of species-groups of Trichoderma: II. production of volatile antibiotics. Transactions of the British Mycological Society, 57, 363–369.
Di Francesco, A., Ugolini, L., Lazzeri, L., & Mari, M. (2015). Production of volatile organic compounds by Aureobasidium pullulans as a potential mechanism of action against postharvest fruit pathogens. Biological Control, 81, 8–14. https://doi.org/10.1016/j.biocontrol.2014.10.004
Di Francesco, A., Milella, F., Mari, M., & Roberti, R. (2017). A preliminary investigation into Aureobasidium pullulans as a potential biocontrol agent against Phytophthora infestans of tomato. Biological Control, 114, 144–149. https://doi.org/10.1016/j.biocontrol.2017.08.010
Di Francesco, A., Calassanzio, M., Ratti, C., Mari, M., Folchi, A., & Baraldi, E. (2018). Molecular characterization of the two postharvest biological control agents Aureobasidium pullulans L1 and L8. Biological Control, 123, 53–59. https://doi.org/10.1016/j.biocontrol.2018.05.005
Di Francesco, A., Di Foggia, M., & Baraldi, E. (2020a). Aureobasidium pullulans volatile organic compounds as alternative postharvest method to control brown rot of stone fruits. Food Microbiology, 87. https://doi.org/10.1016/j.fm.2019.103395
Di Francesco, A., Di Foggia, M., Zajc, J., Gunde-Cimerman, N., & Baraldi, E. (2020b). Study of the efficacy of Aureobasidium strains belonging to three different species: A. pullulans, A. subglaciale and A. melanogenum against Botrytis cinerea of tomato. Annals of Applied Biology, 177, 266–275. https://doi.org/10.1111/aab.12627
Di Francesco, A., Zajc, J., & Stenberg, J. A. (2023). Aureobasidium spp.: Diversity, Versatility, and Agricultural Utility. Horticulturae 2023, 9, 59. https://doi.org/10.3390/horticulturae9010059
Droby, S., Wisniewski, M., Macarisin, D., & Wilson, C. (2009). Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biology and Technology, 52, 137–145. https://doi.org/10.1016/j.postharvbio.2008.11.009
García-Ríos, E., Ramos-Alonso, L., & Guillamón, J. M. (2016). Correlation between low temperature adaptation and oxidative stress in Saccharomyces cerevisiae. Frontiers in Microbiology, 3, 1199. https://doi.org/10.3389/fmicb.2016.01199
Gascuel, O. (1997). BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution, 14, 685–95. https://doi.org/10.1093/oxfordjournals.molbev.a025808
Gostinčar, C., Turk, M., Zajc, J., & Gunde-Cimerman, N. (2019). Fifty Aureobasidium pullulans genomes reveal a recombining polyextremotolerant generalist. Environmental Microbiology, 21, 3638–3652. https://doi.org/10.1111/1462-2920.14693
Gostinčar, C., Ohm, R.A., Kogej, T., Sonjak, S., Turk, M., Zajc, J., Zalar, P., Grube, M., Sun, H., Han, J., Sharma, A., Chiniquy, J., Ngan, C.Y., Lipzen, A., Barry, K., Grigoriev, I. V., & Gunde-Cimerman, N. (2014). Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics, 15. https://doi.org/10.1186/1471-2164-15-549
Gouy, M., Guindon, S., & Gascuel, O. (2010). Sea view version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution, 27, 221–224. https://doi.org/10.1093/molbev/msp259
Huson, D. H., & Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution, 23, 254–267. https://doi.org/10.1093/molbev/msj030
Ippolito, A., El Ghaouth, A., Wilson, C. L., & Wisniewski, M. (2000). Control of postharvest decay of apple fruit by Aureobasidium pullulans and induction of defence responses. Postharvest Biology and Technology, 19, 265–272. https://doi.org/10.1016/S0925-5214(00)00104-6
Janisiewicz, W. J. & Korsten, L. (2002). Biological Control of Postharvest Diseases of Fruits. Annual Review of Phytopathology, 40, 411–441. https://doi.org/10.1146/annurev.phyto.40.120401.130158
Jiang, N., Fan, X., & Tian, C. (2021). Identification and characterization of leaf-inhabiting fungi from castanea plantations in China. Journal of Fungi, 7, 64. https://doi.org/10.3390/jof7010064
Lahlali, R., Aksissou, W., Lyousfi, N., Ezrari, S., Blenzar, A., Tahiri, A., Ennahli, S., Hrustić, J., MacLean, D., & Amiri, S. (2020). Biocontrol activity and putative mechanism of Bacillus amyloliquefaciens (SF14 and SP10), Alcaligenes faecalis ACBC1, and Pantoea agglomerans ACBP1 against brown rot disease of fruit. Microbial Pathogenesis, 139, 103914 https://doi.org/10.1016/j.micpath.2019.103914
Larena, I., Torres, R., de Cal, A., Liñán, M., Melgarejo, P., Domenichini, P., Bellini, A., Mandrin, J. F., Lichou, J., de Eribe, X. O., & Usall, J. (2005). Biological control of postharvest brown rot (Monilinia spp.) of peaches by field applications of Epicoccum nigrum. Biological Control, 32, 305–310. https://doi.org/10.1016/j.biocontrol.2004.10.010
Lee, D., Cho, S., Oh, J., Cho, E., Kwon, S. A. (29021). A novel species of Aureobasidium (Dothioraceae) recovered from Acer pseudosieboldianum in Korea. Journal of Asia-Pacific Biodiversity. 14(4):657–661. https://doi.org/10.1016/j.japb.2021.08.005
Lima, G., Ippolito, A., Nigro, F., & Salerno, M. (1997). Effectiveness of Aureobasidium pullulans and Candida oleophila against postharvest strawberry rots. Postharvest Biology and Technology, 10(2), 169–178. https://doi.org/10.1016/S0925-5214(96)01302-6
Mari, M., Martini, C., Spadoni, A., Rouissi, W., & Bertolini, P. (2012). Biocontrol of apple postharvest decay by Aureobasidium pullulans. Postharvest Biology and Technology, 73, 56–62. https://doi.org/10.1016/j.postharvbio.2012.05.014
Martini, C., Lantos, A., di Francesco, A., Guidarelli, M., D’Aquino, S., & Baraldi, E. (2014). First report of asiatic brown rot caused by Monilinia polystroma on Peach in Italy. Plant Disease, 98, 1585. https://doi.org/10.1094/PDIS-05-14-0551-PDN
Onetto, C. A., Schmidt, S. A.; Roach, M. J., & Borneman, A. R. (2020). Comparative genome analysis proposes three new Aureobasidium species isolated from grape juice. FEMS Yeast Research, 20, foaa052. https://doi.org/10.1093/femsyr/foaa052
Palmieri, D., Ianiri, G., Conte, T., Castoria, R., Lima, G., & De Curtis, F. (2022). Influence of biocontrol and integrated strategies and treatment timing on plum brown rot incidence and fungicide residues in fruits. Agriculture, 12, 1656. https://doi.org/10.3390/agriculture12101656
Pellegrino, C., Gullino, M. L., Garibaldi, A., & Spadaro, D. (2009). First report of brown rot of stone fruit caused by Monilinia fructicola in Italy. Plant Disease, 93, 668. https://doi.org/10.1094/PDIS-93-6-0668A
Pérez-Corral, D. A., Ornelas-Paz, J. D. J., Olivas-Orozco, G. I., Acosta-Muñiz, C. H., Salas-Marina, M. A., Ruiz-Cisneros, M. F., Molina-Corral, F. J., Fernández-Pavía, S. P., & Rios-Velasco, C. (2020). Antagonistic effect of volatile and non-volatile compounds from Streptomyces strains on cultures of several phytopathogenic fungi. Emirate Journal of Food Agriculture, 32, 879–889. https://doi.org/10.9755/ejfa.2020.v32.i12.2222
Petróczy, M., & Palkovics, L. (2009). First report of Monilia polystroma on apple in Hungary. European Journal of Plant Pathology, 125, 343–347. https://doi.org/10.1007/s10658-009-9476-5
Petróczy, M., Szigethy, A., & Palkovics, L. (2012). Monilinia species in Hungary: Morphology, culture characteristics, and molecular analysis. Trees - Structure and Function, 26, 153–164. https://doi.org/10.1007/s00468-011-0622-2
Pinto, C., Custódio, V., Nunes, M., Songy, A., Rabenoelina, F., Courteaux, B., Clément, C., Gomes, A.C., & Fontaine, F. (2018). Understand the potential role of Aureobasidium pullulans, a resident microorganism from grapevine, to prevent the infection caused by Diplodia seriata. Frontiers in Microbiology, 9, 3047.
Rouissi, W., Ugolini, L., Martini, C., Lazzeri, L., & Mari, M. (2013). Control of postharvest fungal pathogens by antifungal compounds from Penicillium expansum. Journal of Food Protection, 76, 1879–1886. https://doi.org/10.4315/0362-028X.JFP-13-072
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D., & Higgins, D. G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7, 539. https://doi.org/10.1038/msb.2011.75
Talibi, I., Boubaker, H., Boudyach, E. H., Aoumar, A. B., & A. (2014). Alternative methods for the control of postharvest citrus diseases. Journal of Applied Microbiology, 117, 1–17. https://doi.org/10.1111/jam.12495
Villarino, M., Egüen, B., Lamarca, N., Segarra, J., Usall, J., Melgarejo, P., & de Cal, A. (2013). Occurrence of Monilinia laxa and M. fructigena after introduction of M. fructicola in peach orchards in Spain. European Journal of Plant Pathology, 137, 835–845. https://doi.org/10.1007/s10658-013-0292-6
Wang, D., Li, Y., Yuan, Y., Chu, D., Cao, J., Sun, G., Ai, Y., Cui, Z., Zhang, Y., Wang, F., & Wang, X. (2022). Identification of non-volatile and volatile organic compounds produced by Bacillus siamensis LZ88 and their antifungal activity against Alternaria alternata. Biological Control, 169, 104901. https://doi.org/10.1016/j.biocontrol.2022.104901
White, T.J., Bruns, T., Lee, S., & Taylor, J., (1990). Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics, in: PCR protocols. A Guide to Methods and Applications, 315–322. https://doi.org/10.1016/b978-0-12-372180-8.50042-1
Zajc, J., Gostinčar, C., Černoša, A., & Gunde-Cimerman, N. (2019). Stress-tolerant yeasts: Opportunistic pathogenicity versus biocontrol potential. Genes, 10. https://doi.org/10.3390/genes10010042
Zalar, P., Gostinčar, C., de Hoog, G. S., Uršič, V., Sudhadham, M., & Gunde-Cimerman, N. (2008). Redefinition of Aureobasidium pullulans and its varieties. Study in Mycology, 61, 21–38. https://doi.org/10.3114/sim.2008.61.02
Zhang, D., Spadaro, D., Garibaldi, A., & Gullino, M. L. (2010). Efficacy of the antagonist Aureobasidium pullulans PL5 against postharvest pathogens of peach, apple and plum and its modes of action. Biological Control, 54, 172–180. https://doi.org/10.1016/j.biocontrol.2010.05.003
Funding
Open access funding provided by Università degli Studi di Udine within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This research did not involve any animal and/or human participants.
Competing interests
The authors declare that they have no conflict of interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cignola, R., Boato, A., Sadallah, A. et al. Molecular characterization of Aureobasidium spp. strains isolated during the cold season. A preliminary efficacy evaluation as novel potential biocontrol agents against postharvest pathogens. Eur J Plant Pathol 167, 221–233 (2023). https://doi.org/10.1007/s10658-023-02696-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-023-02696-x