Abstract
Sex determination is a key developmental event in all organisms. The pathway that regulates sexual fate has been well characterized at the molecular level in the model free-living nematode Caenorhabditis elegans. This study aims to gain a preliminary understanding of sex-determining pathways in a plant-parasitic nematode Meloidogyne incognita, and the extent to which the roles of the sex determination genes are conserved in a hermaphrodite species, C. elegans, and plant-parasitic nematode species, M. incognita. In this study, we targeted two sex-determining orthologues, sdc-1 and tra-1 from M. incognita using RNA interference (RNAi). RNAi was performed by soaking second-stage juveniles of M. incognita in a solution containing dsRNA of either Mi-tra-1or Mi-sdc-1 or both. To determine the effect of RNAi of the target genes, the juveniles treated with the dsRNA were inoculated onto a susceptible cultivar of cowpea grown in a nutrient pouch at 28 °C for 5 weeks. The development of the nematodes was analyzed at different time points during the growth period and compared to untreated controls. Our results showed that neither Mi-sdc-1 nor Mi-tra-1 have a significant role in regulating sexual fate in M. incognita. However, the silencing of Mi-sdc-1 significantly delayed maturity to adult females but did not affect egg production in mature females. In contrast, the downregulation of Mi-tra-1 transcript resulted in a significant reduction in egg production in both single and combinatorial RNAi-treated nematodes. Our results indicate that M. incognita may have adopted a divergent function for Mi-sdc-1 and Mi-tra-1distinct from Caenorhabditis spp. However, Mi-tra-1 might have an essential role in female fecundity in M. incognita and is a promising dsRNA target for root-knot nematode (RKN) management using host-delivered RNAi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sex determination is a fundamental part of reproduction across the entire animal kingdom. However, comparative biology studies have shown that the mechanism that controls sex determination differs considerably among different organisms (Herpin & Schartl, 2008). Invertebrates including worms and insects display a great variety of sex-determining mechanisms (Haag & Doty, 2005). Caenorhabditis elegans, one of the well-known nematode model organisms, has been widely used to systematically characterize sex determination using genetic analysis. Parasitic nematodes, especially plant-parasitic nematodes (PPN) have adopted very distinct life-history from C. elegans, including reproductive strategies. By identifying evolutionary conservation and divergence in the molecular pathway that regulates male and female development in the free-living C. elegans and the plant-parasitic nematode species, it should be possible to understand how complex sex-determining pathways evolve.
PPN is recognized as one of the greatest threats to crops throughout the world. They cause an estimated $80–118 billion dollars per year in damage to crops worldwide (Nicol et al., 2011). Among the different groups of PPN, root-knot nematodes (RKN) are the most economically important group as they can attack and infest a wide range of crops (Elling, 2013; Sasser, 1980). RKN exhibit variable modes of reproduction, meiotic and mitotic parthenogenesis and are extremely difficult to control due to their high reproductive rate, short generation time, and wide host range (Trudgill & Blok, 2001). PPN management using chemical nematicides is highly restricted due to its detrimental effect on humans and the environment. Some of the cultural practices like crop rotation are not always the preferred choice in nematode management (Oka et al., 2000). Developing resistant plants that can suppress nematode growth and development is one of the cost-effective and sustainable methods for managing PPN (Starr et al., 2002). However, the resistance induced by single loci genes can be overcome by some virulent nematode biotypes (Castagnone-Sereno, 2002; Huang et al., 2006). Therefore, developing a new alternative management system for the effective control of RKN species is imperative. To develop nematode resistance targeting nematode fecundity to reduce the multiplication of RKN in host plants, an insight into the sex determination pathway in nematodes might be an advantage. Our knowledge of the sex determination mechanism in plant-parasitic nematodes is very limited. This study aims to understand if some of the known C. elegans sex-determination homologs have a similar functional role in determining sexual fate in the root-knot nematode, M. incognita.
Comparative studies of sex determination based on the Caenorhabditis elegans model have revealed both rapidly evolving and well-conserved features among different species of nematodes (Haag, 2005). Decades of research with C. elegans have shown that sexual development in both the somatic tissues and the germline is regulated by a signal transduction pathway. The hermaphrodite females and males of C. elegans have different sex chromosome numbers i.e. XX/XO respectively and the sex is primarily determined by the ratio between the number of X chromosomes and sets of autosomes (Zanetti & Puoti, 2013). The remaining part of the sex determination pathway is defined by a cascade of negative regulatory interactions that alternately activate or repress male- or female-specific genes. More specifically, SDC (Sex determination and Dosage Compensation defect) proteins negatively regulate the production of a hormone, HER-1 (human epidermal growth factor receptor-1), that causes cells throughout the body to adopt male fates by repressing tra-2. In XX animals, the absence of HER-1 promotes the expression of tra-2, which in turn negatively regulates the male promoting genes, feminization-1 (fem-1), feminization-2 (fem-2), feminization-3 (fem-3) (Zarkower, 2006). The repression of FEM proteins results in the activation of the terminal target of this pathway, tra-1, a transcription factor, which promotes the female fate in XX animals by regulating downstream targets required for male development, fog-1, and fog-3 (Ellis & Lin, 2014).
However, some groups of nematodes do not rely only on genetic sex determination but also depend on environmental sex determination (Christie, 1929; Harlos et al., 1980). Previous studies have shown that environmental factors can influence the sexual fate in M. incognita (Triantaphyllou, 1973). Under suitable environmental conditions and hosts, most offspring become female at maturity. Sex reversal of female to male and intersex phase also has been reported (Papadopoulou & Traintaphyllou, 1982). M. incognita typically reproduces by mitotic parthenogenesis (Abad et al., 2008) and genetic contribution to the offspring is fully dependent on the females. Comparative genome analysis by Abad et al. (2008) identified M. incognita homologs of at least one member of each step of the C. elegans sex determination cascade, including sdc-1 from the dosage compensation pathway, tra-1, tra-3, and fem-2 from the sex determination pathway itself. The presence of these homologs in a mitotic parthenogenetic nematode which displays an environmental influence on sex determination, attracted our attention to study further.
Decades of studies in C. elegans have elucidated the complex sex determination pathway with systematic characterization and genetic analysis. The study on mutant alleles of X-linked gene sdc-1 has shown that it is required for the coordinate control of the sex determination and dosage compensation processes in C. elegans (Villeneuve & Meyer, 1987). In XX animals, these sdc-1 mutations resulted in both masculinization and increased levels of X-linked gene expression, causing a shift towards male fate (Yonker & Meyer, 2003). Similarly, in C. elegans, tra-1 loss-of-function XX mutant animals are somatically transformed into males (Hodgkin & Brenner, 1977). The current study is an attempt to answer the following questions: What is the functional role of sdc-1 in a parthenogenetic nematode species? Is there any genetic contribution from female promoting genes such as tra-1 in determining the female fate in M. incognita? Do they have any divergent functional roles in M. incognita adopted during the evolution of parasitic nematodes?
Secondly, we explore the possibility of using RNAi for the functional characterization of genes involved in the sex determination of M. incognita. Considering the population dynamics and crop damage caused by M. incognita, the factors that determine the sex ratio is of fundamental importance. Additionally, the extent of damage caused by M. incognita is directly proportional to the female population because females require more food for increased reproduction. Therefore, it would be advantageous if we could shift the sex ratio of M. incognita towards male direction by any means. In this study, we tested the possibility of reversing sex from female to male or reducing female fecundity by knocking down the C. elegans homologs of female promoting genes, Mi-tra-1 and Mi-sdc-1 in M. incognita. Since the application of current advanced gene-editing technology such as CRISPR/cas9 is currently not possible in parthenogenetic sedentary species of PPN, we chose RNAi to knock down the expression of the target genes. If the manipulation of sexual fate is possible by RNAi of Mi-tra-1 or Mi-sdc-1, they would be a potential target to introduce M. incognita resistance in susceptible crops via in planta RNAi.
Materials and methods
Nematode cultures
A homogeneous population of M. incognita was obtained from the University of Florida Nematode Assay Lab stock cultures that were maintained under greenhouse conditions on tomato (Solanum lycopersicum L. cv. Rutgers). The identity of the culture was confirmed by PCR. Briefly, DNA was extracted from eggs using the Proteinase K method (Blin & Stafford, 1976) and PCR was done using M. incognita specific primers followed by gel electrophoresis. Eggs were extracted from galled tomato roots using 20 ml of 10% sodium hypochloride (NaOCl) in 980 ml water and incubated at 28 °C for 3 days in a hatching disc. Freshly hatched juveniles were collected by a modified Baermann dish technique. The juvenile suspension was adjusted to 1000 J2/ml of water and used as inoculum to inoculate in each pouch.
Selection of target gene and primer design
Sex determining genes of C. elegans were compared with the whole genome sequence of M. incognita by Abad et al., 2008. Based on the results of comparative analysis, we selected orthologs of two female promoting genes, downstream (tra-1) and upstream (sdc-1) genes in C. elegans as our target genes for studying sex determination pathway in M. incognita. The protein sequences of tra-1 and sdc-1 of C. elegans retrieved from WormBase (Howe et al., 2017) were used as queries to perform blastP in addition to tblastn against the M. incognita genome at WormBase Parasite database (https://parasite.wormbase.org/Multi/Tools/Blast) to retrieve homologs of tra-1 and sdc-1 in M. incognita (Mi-tra-1 and Mi-sdc-1). To check if the resulting top hit was a true homolog of the C. elegans gene, the resulting coding and protein sequence was compared with (blastx, E < 1e-5) C. elegans proteins at the Wormbase database. A different set of primers for each gene was designed to amplify the target exon region of best-hit nucleotide sequences (Table 1).
RNA extraction and cloning of Mi-tra-1 and Mi-sdc-1.
Total RNA was extracted from eggs and second-stage juveniles of M. incognita using the Trizol method according to the manufacture’s protocol (Invitrogen, Thermo Fisher Scientific, CA). The first-strand cDNA was synthesized using 1000 ng of RNA, 20 mM dNTP, 100 μM oligo dT primers, 400 units of Reverse Transcriptase (SuperScript® IV RT, Invitrogen Thermo Fisher Scientific, CA), 80 units of Ribonuclease inhibitor, and 0.2 M of DTT. After RNA extraction, it was treated with DNAse to avoid contamination. A 228 bp and 462 bp was amplified using Mi-tra1F1 and Mi-tra1R1/R2 respectively targeting Mi-tra-1 whereas a 240-bp and 298 bp of Mi-sdc-1 were amplified using primers Mi-sdc1F1 and Mi-sdc1R1/R2 respectively targeting Mi-sdc-1 from the cDNA of M. incognita (Table 1). The PCR conditions for Mi-tra-1 and Mi-sdc-1 were: 95 ͦ C for 15 min followed by 40 cycles of 95 ͦ C for 1 min, 50 ͦ C for 45 s and 72 ͦ C for 1 min, followed by incubation at 72 ͦ C for 7 min. Amplified fragments were gel purified using Qiagen QIAquick Gel Extraction Kit (Qiagen Inc., CA) following the manufacture’s protocol and were sent to the University of Florida’s campus-wide Interdisciplinary Center for Biotechnology Research (ICBR) DNA Sequencing Facility (Gainesville, FL) for Sanger sequencing.
Confirmed amplified fragments were ligated to a pDrive cloning vector and introduced into a Qiagen EZ competent cell followed by transformation of the competent cell using the manufacturer’s protocol (QIAGEN® PCR Cloning Handbook, CA). Recombinant plasmids were then purified using Zyppy™ Plasmid Miniprep Kit (Zymo Research, CA) following the manufacturer’s protocols. Expression of Mi-tra-1 and Mi-sdc-1 were assessed for all life stages of the nematode (egg, J2, J3, J4, early adult, and mature female). For the expression study, total RNA was extracted from all life stages. Briefly, about 25 nematodes were picked manually from the nematode infected tomato roots 8, 12, 16, 23, and 30 days after inoculation of J2 onto the plant roots. RNA extraction was done by using the Trizol method (Invitrogen, Thermo Fisher Scientific Carlsbad, CA, USA) according to the manufacture’s protocol followed by cDNA synthesis and PCR for 35 cycles as described previously. The housekeeping gene, Mi-actin was used as a control and amplified using Mi-actin F and Mi-actin R primer (Table 1) (Duarte et al., 2016).
Synthesis of double-stranded RNA
The desired template of cDNA containing Mi-tra-1 (228 bp) and Mi-sdc-1 (240 bp) genes from J2 was amplified by PCR to generate dsRNA of the respective genes. The Mi-tra-1 templet of length 228 bp and Mi-sdc-1 of 240 bp were selected to assure uniformity during dsRNA synthesis. The standard condition for PCR was 95 ͦ C for 10 min followed by 35 cycles of 95 ͦ C for 1 min, 50 ͦ C for 45 s and 72 ͦ C for 1 min, which was followed by incubation at 72 ͦ C for 7 min. The respective primers with the T7 promoter sequence incorporated at the 5′ end of either the sense or antisense strand were used for PCR amplification (Table 1). PCR products were transcribed and purified using Ambion MEGAscript® RNAi Kit (Ambion, UK) according to the manufacturer’s protocol. The double-stranded RNA was made by incubating equimolar amounts of sense and antisense strands in boiling water for 5 min, followed by 1 h at room temperature and subsequent treatment with DNAse to remove the template. The dsRNA was quantified spectrophotometrically and was checked on 1.5% agarose gel. To check dsRNA toxicity due to exogenous dsRNA supply, the dsRNA of Green Fluorescent Protein (GFP) was used as a control as it does not have a direct impact on sex determination in M. incognita. The use of dsRNA of gfp helped to assess the non-specific effect of dsRNA on nematode sex determination. The sense and antisense strand of gfp with its promoter region was amplified as described previously using the primers with the T7 promoter sequence incorporated at the 5’end of either sense or antisense strand (Table 1). The dsRNA of gfp was synthesized as described for Mi-tra-1 and Mi-sdc-1.
In vitro RNAi of M. incognita and analysis of mRNA levels by semi-quantitative RT-PCR
For the soaking assay, approximately 7000 J2 of freshly collected M. incognita were incubated for 24 h in 200 μl of a solution containing 100 μl of dsRNA (2 μg/μl), 50 mM octopamine (Sigma- Aldrich, MO), 3 mM spermidine (Sigma- Aldrich, MO), 2 μl of 5% gelatin (Sigma- Aldrich, MO) and 80 μl of distilled water with nematodes for each of the three treatments, i.e., Mi-tra-1, Mi-sdc-1 and gfp dsRNA keeping the final concentration of dsRNA as 1 μg/μl. For negative control, the same solution without dsRNA was used. For a second experiment, all the treatments were the same as the first with additional combinatorial dsRNA treatments consisting of dsRNAs of Mi-tra-1 and Mi-sdc-1. For the preparation of combinatorial dsRNA, the final concentration of dsRNA was kept as 1 μg/μL which was similar to the individual dsRNA treatments. The addition of octapamine, gelatin, and spermidine into dsRNA solution improves the efficacy of RNAi uptake by nematode as described by Sukno et al., 2007. The nematodes were incubated at 28 ͦ C for 24 h in 1.5 ml micro-centrifuge Eppendorf tubes. After incubation, the nematodes were washed thoroughly with distilled water by centrifugation at 4000 rpm for 1 min. The washing was repeated three times. After washing approximately 400 nematodes from each treatment were used for total RNA extraction using Trizol reagent (Invitrogen, Thermo Fisher Scientific, CA) according to the manufacturer’s instructions. Equal amounts of RNA from each treatment were used in cDNA synthesis as described in the above section. Prepared RNA was treated with DNAse to remove genomic DNA. A similar process was repeated 48, 72, and 96 h after recovery.
Qualitative and quantitative characterization of knock-down phenotypes
Double-stranded RNA-treated nematodes were washed with sterile water and monitored using a microscope to check for effects on mortality due to dsRNA toxicity. 500 μl of sterile water containing approximately 500 nematodes from each dsRNA treatment were inoculated onto the cowpea seedlings. Nematodes were distributed throughout the roots of the plant using a pipet. Treated plants were arranged in Random Complete Block Design (RCBD) with 12 replicates from each treatment. Fifteen days after inoculation, six pouches from each treatment were harvested and the roots were stained using the sodium-hypochlorite-acid-fuschin method (Byrd et al., 1983). The number of nematodes in each root with each developmental stage was recorded. Each developmental stage was determined based on the morphological characteristics of Abad et al., 2008. Thirty days after inoculation, the remaining six plants per treatment were harvested and stained as described previously. The number of females with eggs and without eggs was counted from each plant’s roots. Pink stained root galls were counted as the females with egg masses and the unstained galls were dissected to check the presence of females within the gall. The females without egg masses from the unstained galls were dissected to examine the gonads. Data analysis was done using one-way ANOVA with multiple mean comparisons between treatments using Tukey’s test (P = 0.05). All experiment was repeated once.
Preparation of plant for inoculation
Cowpea (Vigna unguiculata. cv. Whippoorwill) was used for inoculation assay. Cowpea seeds were sterilized with 70% ethanol and 0.1% bleach followed by incubation inside the incubator for 24 h at 28 ͦ C to enhance germination. After germination, seeds were transferred to CYG seed growth pouches (Mega International, Minneapolis, MN, USA) and kept inside a growth chamber at 28 ͦ C for 15 days to allow for root development. For the quantification of knock-down phenotype, dsRNA treated J2 were inoculated in the cowpea plants. After inoculation, the cowpea plants were laid on the bench for 24 h in dark to ensure nematode penetration and transferred to an upright position inside the incubator (Fig. 1).
Results
A high degree of sequence conservation of tra-1 and sdc-1 orthologs among different root-knot nematodes
The sequence obtained from amplification of cDNA of M. incognita using primer Mi-tra-1F1 and Mi-tra1R2 and subsequent cloning followed by Sanger sequencing was used as a query (228 bp) to perform blastx against the genomic database from WormBase Parasite. The blast result was100% similar to the Zinc Finger protein-like sequences (Minc3s00848g18057) from M. incognita (PRJEB8714). Similarly, amplicon obtained from PCR using Mi-sdc-1F1 and Mi-sdc1R1 was used as a query (240 bp) to perform blastx against M. incognita genome database. The blast result indicated 100% similarity with the protein sequence (Minc3s00642g15517) from M. incognita (PRJEB8714). The reverse BLAST (blasx, E < 1e-5) against the C. elegans genome database confirmed that the identified gene from M. incognita are true orthologs of C. elegans genes, tra-1 and sdc-1. The amplified fragment (228 bp) from Mi-tra-1 was 100% similar to the ortholog sequences from M. javanica, M. arenaria, and M. floridensis over the entire sequence. A BLAST search for conserved domain from the retrieved protein sequences of (Mi-tra-1 and Mi-sdc-1) from M. incognita and other RKN species showed the presence of a Cys2His2 (C2H2)-type zinc finger site as in tra-1 homolog in C. elegans. Similarly, the selected amplicon (240 bp) from Mi-sdc-1, which was used for dsRNA synthesis showed 100% similarity with the orthologs of sdc-1 from M. arenaria and M. javanica while it was 97.1% and 96.7% similar for M. enterolobii and M. floridensis respectively. However, M. hapla, an amphimictic root-knot nematode, showed only 87.9% similarity with the selected sequence of Mi-sdc-1. Thus, the selected nucleotide sequences used in this study for dsRNA synthesis targeting Mi-tra-1 and Mi-sdc-1 is highly conserved among different root-knot nematode species and also the protein alignment of both tra-1 and sdc-1 orthologs from M. incognita and other RKN species confirmed that these two genes are highly conserved among different RKN species (Fig. 2 A and B).
Protein alignment of Mi-tra-1 and Mi-sdc-1 with its orthologs from closely related root-knot nematode (RKN) species and C. elegans. (A) Protein alignment of Mi-tra-1 showing high sequence conservation among the RKN species and C. elegans. (B). Protein alignment of Mi-sdc-1 showing high sequence conservation among the RKN species C. elegans. The highlighted region shows the sequence region used for constructing dsRNA for the study
Mi-tra-1 and mi-sdc-1 can be efficiently silenced by RNAi
Semi-quantitative RT-PCR on cDNA was done on different life stages of M. incognita to check the expression level of Mi-tra-1, Mi-sdc-1, and the housekeeping gene actin. The transcripts of both Mi-tra-1 and Mi-sdc-1 were detected in all developmental stages including eggs, J2, J3, J4, early adult (EA) or young adult, and matured female (MF). However, the expression of Mi-tra-1 was relatively low in all stages as compared to Mi-sdc-1 and the transcription level of Mi-tra-1 was higher in matured females than in other stages (Fig. 3).
Based on the results from the semi-quantitative RT-PCR, a significant and specific reduction in transcript level of target genes was observed in the nematodes that were incubated in the corresponding dsRNA solution for 24 h as compared to the controls, gfp RNAi, and control (Fig. 4). Freshly hatched juveniles soaked in the dsRNA solutions were also subjected to further investigation to check the durability of gene knock-down. Analysis of RT-PCR revealed that the transcript level of Mi-tra-1 and Mi-sdc-1 were significantly reduced after 24 h, 48 h, 72 h, and 96 h (Fig. 4). However, in the case of Mi-sdc-1 RNAi, a gradual recovery of transcript level was observed after 24 h while a strong reduction in expression was persistent in Mi-tra-1RNAi even 96 h after recovery (Fig. 4). The recovered J2s monitored under a microscope showed that most of the nematodes that were soaked for 24 h in dsRNA of Mi-tra-1, Mi-sdc-1 as well as gfp showed normal sinusoidal movement as observed in the control solution (Fig. 5). This suggests that there was no significant non-specific RNAi effect on nematode movement due to soaking in dsRNA and changes in nematode phenotypes during the later stage of nematode is gene-specific and the result of targeted silencing.
Meloidogyne incognita J2 RNAi phenotype was monitored under a microscope after treating for 24 h with dsRNA of Mi-tra-1, Mi-sdc-1, GFP, and control in the first experiment and additional dsRNA treatment of Mi-tra-1 combined with Mi-sdc-1 in the second experiment. (A) Nematodes treated with control treatment. (B) Nematodes treated with GFP dsRNA. (C) Nematodes treated with Mi-sdc-1 dsRNA. (D) Nematodes treated with Mi-tra-1 dsRNA, and (E) Nematode treated with combinatorial dsRNA of Mi-tra-1 and Mi-sdc-1. All the observation is taken in 10x magnification (AxioCam MRc 5, Zeiss)
RNAi of mi-sdc-1 delayed development of young female from J3 and J4 stage while RNAi of mi-tra-1 caused reduced fecundity in mature females
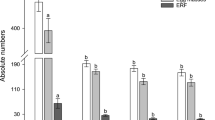
On the 15th day after inoculation, 91% of the nematode population was at the third and fourth juvenile stage and 9% of the population at the early female stage in the treatment with the dsRNA of Mi-sdc-1, which was significantly different from all other treatments (P ≤ 0.05). In the case of Mi-tra-1 RNAi treatment, 72% of nematodes were at the third and fourth juvenile development stages and 28% of nematodes at the early female stage. This result was not significantly different from control (68% third and fourth stage juveniles and 32% early female stage), and gfp dsRNA treatments (70% third and fourth stage juveniles and 30% of early female) (Fig. 6).
Effects of Mi-tra-1 and Mi-sdc-1 RNAi treatment on Meloidogyne incognita after 15 days. A significantly high percentage (91%) of the nematode population was found at the third and fourth juvenile stage (J3 + J4) following treatment with the dsRNA of Mi-sdc-1 compared to that of early females (EF), which was different (P ≤ 0.05) from all other treatments. Error bars indicate standard error
On the 35th day after inoculation, roots were stained and females with egg masses and without egg masses were counted (Fig. 7). The results showed that among the nematodes treated with Mi-tra-1 dsRNA, 65% of females were without eggs and 35% were females with eggs, which was significantly different from other treatments (P ≤ 0.05). Nematodes treated with Mi-sdc-1 dsRNA had 12% females without eggs and 88% of females with eggs, which was not significantly different from gfp RNAi (87% females with eggs and 13% females without eggs) or control (96% females with eggs and 4.0% of females without eggs) (Fig. 8A). Combinatorial dsRNA treatment (with both Mi-tra-1 and Mi-sdc-1) resulted in 84% females without eggs and 16% females with eggs, which is significantly different from single dsRNA treatment of Mi-sdc-1 or Mi-tra-1 and gfp (P ≤ 0.05). Similarly, Mi-tra-1 resulted in 72% females without eggs and 28% females with eggs which were significantly different from the control treatment. However, Mi-sdc-1 resulted in 80% of females with eggs, which was not significantly different from gfp dsRNA treatment (Fig. 8B).
Acid-fuschin stained cowpea roots after 35 days post-inoculation with Meloidogyne incognita J2 treated with dsRNAs. (A). Treatment with dsRNA of Mi-sdc-1. Fully mature females with egg masses were observed. The egg masses are stained pink. (B) Control was treated with a soaking solution without dsRNA. (C). Treatment with dsRNA of Mi-tra-1, which shows a reduced number of females without egg masses (lack of pink stained egg masses). The unstained galls were dissected to check the presence of the females inside the galls and the females without egg masses also lacked eggs within the female body. The galls are enlarged and indicated by the red arrow
(A). Effects of Mi-tra-1 and Mi-sdc-1 RNAi treatment on Meloidogyne incognita after 35 days. Grey bars indicate the percentage of females without eggs and black bars indicate females with eggs. Error bars indicate standard error. (B). Comparison of single RNAi with single dsRNA of Mi-tra-1 and Mi-sdc-1and the combinatorial RNAi with the dsRNA of both target genes together on the phenotype of M. incognita after 35 days. Error bars indicate standard error
Discussion
In most animal species, sex determination is the fundamental step that it takes to become male or female. However, the mechanism that determines sexual determination is highly diverse. This study is to gain an initial understanding of the role of C. elegans sex determination homologs, Mi-tra-1, and Mi-sdc-1, in defining the sexual fate in M. incognita. In C. elegans, the studies have shown that the mutations in the gene, sdc-1 disrupt both sex determination and dosage compensation in XX animals and lack of sdc-1 function causes an incomplete transformation of XX animals to males. In contrast, the reduced expression of Mi-sdc-1 in M. incognita did not result in somatic change towards the male phenotype. However, in our study, we observed that 91% of nematodes were at the third or fourth juvenile stage, which was significantly different from the control treatments at 15 days post-inoculation of nematodes with dsRNA of Mi-sdc-1. However, 35 days post-inoculation, there was no significant difference in the number of mature females with eggs as compared to the control treatment. This data indicates that reduced expression of Mi- sdc-1 resulted in a significant delay in the development from the J3/J4 stage to the female stage but did not arrest further development to fully matured females.
The studies in C. elegans have shown that sdc-1 acts during the first half of embryogenesis, especially at one or two-celled embryos, in determining somatic sexual phenotype, long before sexual differentiation takes place and expressed in all life stages (Villeneuve & Meyer, 1987). Although Mi-sdc-1 is expressed from embryo to the adult female stage in M. incognita, its role in an early step in the regulatory hierarchy controlling the choice of sexual fate remains unclear from our data since we did not find a significant shift toward male fate. However, the silencing effect of Mi-sdc-1 did not persist after recovering the nematodes from dsRNA treatment. A slow recovery in the gene expression of Mi- sdc-1 was observed 96 h after the withdrawal of dsRNA (Fig. 4). Additionally, we did not quantify the expression level of Mi-sdc-1 in the treated nematodes during its development within the plant. The mi-sdc-1 expression might be recovered completely after 35 days of infection, and thus did not observe a significant phenotypic effect in the nematodes. Additionally, the homology search against updated M. incognita whole-genome did not find true homologs of other genes in SDC assembly, sdc-2 and sdc-3 as well as her-1, which is negatively regulated by the SDC proteins to promote the sexual fate toward females in C. elegans. This gives the notion that the initiation of sex determination cascade mediated by SDC proteins in the X chromosome dosage compensation pathway might not be functionally conserved in M. incognita as in C. elegans.
Similarly, the RNAi phenotype of Mi-tra-1 was very different from the mutant phenotype of tra-1 in C. elegans. A complete loss of function of tra-1in C. elegans transforms XX animals to pseudo males with male soma but the germline produces both sperm and oocytes (Schedl et al., 1989). Thus tra-1 is essential for somatic female development in C. elegans. In contrast, we did not find a significant increase in male nematodes after treating the nematodes with dsRNA of Mi-tra-1 but a significantly high number of females were without eggs (Fig. 8) in the treated nematodes as compared to control treatments. Though RNAi of Mi-tra-1-RNAi did not induce any somatic change in the female, it might be possible that the development of the female germline could get affected and resulted in low egg production. However, we could not confirm if there was an abnormality in gonads or germline development in the treated nematodes due to poor visibility of germ cells under DIC microscopy. Although tra-1 is mostly known as a master regulator of sex determination in Caenorhabditis species to promote female fate, it is a fly and vertebrate homologs that regulate embryonic patterning and cell proliferation (Ingham & McMahon, 2001). Additionally, well-documented studies have shown that TRA-1 is also a key regulator in the formation of somatic gonadal precursor cells (SGPs) (Mathies et al., 2004), and gonadal cell proliferation (Chang et al., 2004) in both XX and XO animals in C. elegans. Therefore, tra-1 might have a prominent role in female gonadogenesis in M. incognita. Based on the BLAST analysis using the updated whole genome database, M. incognita lacks true homologs of upstream regulators of tra-1 in C. elegans such as tra-2, fem-1, and fem-3. The FEM genes, fem-1, fem2, and fem-3 are the key male-promoting genes by negatively regulating tra-1 to promote spermatogenesis in C. elegans. However, M. incognita has a homolog for fem-2 alone and its functional role yet to be determined in this apomictic nematode, which can reproduce asexually without sperm. In C. elegans, the binding of tra-2 with FEM genes is necessary to activate tra-1 to promote female fate (Wang & Kimble, 2001). The absence of tra-2 and other FEM homologs in M. incognita, and our results showing that knocking down tra-1 alone is not enough to generate masculinization of females suggests that Mi-tra-1 might not have a significant role in regulating somatic sex determination in M. incognita. However, the most recent M. incognita assembly shows a hybrid origin with two divergent genomic copies, and several copies of one gene may present within the genome (Szitenberg et al., 2017). Based on our blast search against the updated M. incognita genome database showed that Mi-sdc-1 shares significantly high homology with the other 3 paralogs whereas Mi-tra-1 showed less homology with its paralogous genes. This implies that there might be a functional overlap among these genes and further characterization of these genes is required to understand the regulation of sex determination in M. incognita.
In this study, we tested the impact of RNAi of single soaking with dsRNA of either Mi-tra-1 or Mi-sdc-1 alone as well as the soaking with the combination of the two. In the case of a single soaking experiment, we observed two different RNAi phenotypes, delayed development after 15 days in Mi-sdc-1 RNAi, and reduced fecundity after 35 days in Mi-tra-1 RNAi. In the case of combinatorial RNAi, we did not observe delayed phenotype after 15 days (data not shown), but a significant reduction in fecundity after 35 days was observed as in the case of single RNAi of Mi-tra-1. This can be explained based on factors that influence the dsRNA dose required for efficient RNAi in M. incognita. In both single and combinatorial RNAi, we used total dsRNA at a maximum concentration of 1 μg/μl in the soaking solution because a non-specific RNAi effect was observed in nematodes soaked with dsRNA of gfp at a higher dosage beyond 1 μg/μl. Hence, in the combinatorial soaking, the concentration of dsRNA of each Mi-tra-1 and Mi-sdc-1 in the total solution was 500 ng/ μl which was 50% of dsRNA dose used in a single soaking. Although dsRNA at 500 ng/ μl was good enough to induce a stronger Mi-tra-1 RNAi phenotype in the combinatorial soaking, it did not show RNAi phenotype of Mi-sdc-1. This discrepancy in knockdown effect at lower dose can be due to several factors including differing spatial expression patterns level, and turnover rate of the transcript. In our observation, the expression of Mi-sdc-1 transcript was higher than Mi-tra-1 in every development stage and might require a higher dose of dsRNA to induce an efficient knockdown effect in the nematode. Current advances in genetic modification of plant-parasitic nematodes by delivering macromolecules to male germline (Kranse et al., 2021) gives a promising approach to functionally characterize C. elegans homologs of sex determination pathway in RKN species. In contrast, RNAi-mediated knockdown is less efficient to access an accurate scorable phenotype.
In addition to gaining preliminary insight on the sex determination pathway in M. incognita, our in vitro RNAi results shows that Mi-tra-1 could be a promising target for further study to develop host-delivered RNAi (HD-RNAi)-mediated resistance against M. incognita. The efficiency of HD RNAi relies on the knowledge of appropriate nematode genetic targets for RNAi. Although many studies have identified various nematode genes required for parasitism and housekeeping genes involved in nematode development by in vitro RNAi (Jaouannet et al., 2013; Sindhu et al., 2009; Yadav et al., 2006). To our best knowledge, this is the first report on characterizing C. elegans sex determination homologs in a plant-parasitic nematode. One of the basic principles of nematode management is to adopt a strategy that reduces the rate of nematode population increase. In root-knot nematodes, the females directly determine the extent of damage it causes to the crops. In this study, we aimed to alter the sex ratio toward males by knocking down female-promoting genes. However, the downregulation of Mi-tra-1 did not increase the percentage of males, but it did affect the rate of female reproduction. Therefore, the rate of nematode increase in subsequent generations can be reduced if transgenic plants were developed that continuously generate dsRNA or siRNA of Mi-tra-1. Due to high conservation among different RKN species, Mi-tra-1 might be a suitable target for HD-RNAi for broader management of different RKNs including virulent species such as M. floridensis.
References
Abad, P., Gouzy, J., Aury, J.-M., Castagnone-Sereno, P., Danchin, E. G. J., Deleury, E., Perfus-Barbeoch, L., Anthouard, V., Artiguenave, F., Blok, V. C., Caillaud, M. C., Coutinho, P. M., Dasilva, C., de Luca, F., Deau, F., Esquibet, M., Flutre, T., Goldstone, J. V., Hamamouch, N., Hewezi, T., Jaillon, O., Jubin, C., Leonetti, P., Magliano, M., Maier, T. R., Markov, G. V., McVeigh, P., Pesole, G., Poulain, J., Robinson-Rechavi, M., Sallet, E., Ségurens, B., Steinbach, D., Tytgat, T., Ugarte, E., van Ghelder, C., Veronico, P., Baum, T. J., Blaxter, M., Bleve-Zacheo, T., Davis, E. L., Ewbank, J. J., Favery, B., Grenier, E., Henrissat, B., Jones, J. T., Laudet, V., Maule, A. G., Quesneville, H., Rosso, M. N., Schiex, T., Smant, G., Weissenbach, J., & Wincker, P. (2008). Genome sequence of the metazoan plant parasitic nematode Meloidogyne incognita. Nature Biotechnology, 26(8), 909–915. https://doi.org/10.1038/nbt.1482.
Blin, N., & Stafford, D. W. (1976). A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Research, 3(9), 2303–2308 http://www.ncbi.nlm.nih.gov/pubmed/987581.
Byrd, D. W., Kirkpatrick, T., Barker, K. R., & Barker, K. R. (1983). An improved technique for clearing and staining plant tissues for detection of nematodes. Journal of Nematology, 15(1), 142–143 http://www.ncbi.nlm.nih.gov/pubmed/19295781.
Castagnone-Sereno, P. (2002). Genetic variability of nematodes: A threat to the durability of plant resistance genes? Euphytica, 124(2), 193–199. https://doi.org/10.1023/A:1015682500495.
Chang, W., Tilmann, C., Thoemke, K., Markussen, F. H., Mathies, L. D., Kimble, J., & Zarkower, D. (2004). A forkhead protein controls sexual identity of the C. elegans male somatic gonad. Development. The Company of Biologists Ltd. https://doi.org/10.1242/dev.01012.
Christie, J. R. (1929). Some observations on sex in the Mermithidae. Journal of Experimental Zoology, 53(1), 59–76. https://doi.org/10.1002/jez.1400530106.
Duarte, A., Maleita, C., Tiago, I., Curtis, R., & Abrantes, I. (2016). Molecular characterization of putative parasitism genes in the plant-parasitic nematode Meloidogyne hispanica. Journal of Helminthology, 90(01), 28–38. https://doi.org/10.1017/S0022149X1400073X.
Elling, A. A. (2013). Major emerging problems with minor Meloidogyne species. Phytopathology, 103(11), 1092–1102. https://doi.org/10.1094/PHYTO-01-13-0019-RVW.
Ellis, R. E., & Lin, S. Y. (2014). The evolutionary origins and consequences of self-fertility in nematodes. F1000Prime Reports, 6(62). https://doi.org/10.12703/P6-62.
Haag, E. S. (2005). The evolution of nematode sex determination: C. elegans as a reference point for comparative biology. WormBook. https://doi.org/10.1895/wormbook.1.120.1.
Haag, E. S., & Doty, A. V. (2005). Sex determination across evolution: Connecting the dots. PLoS Biology, 3(1), e21. https://doi.org/10.1371/journal.pbio.0030021.
Harlos, J., Brust, R. A., & Galloway, T. D. (1980). Observations on a nematode parasite of Aedes vexans (Diptera:Culicidae) in Manitoba. Canadian Journal of Zoology, 58(2), 215–220. https://doi.org/10.1139/z80-024.
Herpin, A., & Schartl, M. (2008). Regulatory putsches create new ways of determining sexual development. EMBO Reports, 9(10), 966–968. https://doi.org/10.1038/embor.2008.182.
Hodgkin, J. A., & Brenner, S. (1977). Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics, 86(2), 275–287.
Howe, K. L., Bolt, B. J., Shafie, M., Kersey, P., & Berriman, M. (2017). WormBase ParaSite − a comprehensive resource for helminth genomics. Molecular and Biochemical Parasitology, 215, 2–10. https://doi.org/10.1016/j.molbiopara.2016.11.005.
Huang, G., Allen, R., Davis, E. L., Baum, T. J., & Hussey, R. S. (2006). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proceedings of the National Academy of Sciences of the United States of America, 103(39), 14302–14306. https://doi.org/10.1073/pnas.0604698103.
Ingham, P. W., & McMahon, A. P. (2001). Hedgehog signaling in animal development: Paradigms and principles. Genes and Development. Cold Spring Harbor Laboratory Press. https://doi.org/10.1101/gad.938601.
Jaouannet, M., Magliano, M., Arguel, M. J., Gourgues, M., Evangelisti, E., Abad, P., & Rosso, M. N. (2013). The root-knot nematode calreticulin mi-CRT is a key effector in plant defense suppression. Molecular Plant-Microbe Interactions, 26(1), 97–105. https://doi.org/10.1094/MPMI-05-12-0130-R.
Kranse, O., Beasley, H., Adams, S., Pires-Da Silva, A., Bell, C., Lilley, C. J., et al. (2021). Toward genetic modification of plant-parasitic nematodes: Delivery of macromolecules to adults and expression of exogenous mRNA in second stage juveniles. G3: Genes, genomes, genetics, 11(2). https://doi.org/10.1093/g3journal/jkaa058.
Mathies, L. D., Schvarzstein, M., Morphy, K. M., Blelloch, R., Spence, A. M., & Kimble, J. (2004). TRA-1/GLI controls development of somatic gonadal precursors in C. elegans. Development, 131(17), 4333–4343. https://doi.org/10.1242/dev.01288.
Nicol, J. M., Turner, S. J., Coyne, D. L., Nijs, L. den, Hockland, S., & Maafi, Z. T. (2011). Current nematode threats to world agriculture. In genomics and molecular genetics of plant-nematode interactions (pp. 21–43). Springer Netherlands. https://doi.org/10.1007/978-94-007-0434-3_2.
Oka, Y., Koltai, H., Bar-Eyal, M., Mor, M., Sharon, E., Chet, I., & Spiegel, Y. (2000). New strategies for the control of plant-parasitic nematodes. Pest Management Science, 56(11), 983–988. https://doi.org/10.1002/1526-4998(200011)56:11<983::AID-PS233>3.0.CO;2-X.
Papadopoulou, J., & Traintaphyllou, A. C. (1982). Sex differentiation in Meloidogyne incognita and anatomical evidence of sex reversal. Journal of Nematology, 14(4), 549–566 http://www.ncbi.nlm.nih.gov/pubmed/19295752.
Sasser, J. N. (1980). Root-knot nematodes: A global menace to crop production. Plant Disease, 64(1), 36–41 https://www.cabdirect.org/cabdirect/abstract/19790866155.
Schedl, T., Graham, P. L., Barton, M. K., & Kimble, J. (1989). Analysis of the role of tra-1 in germline sex determination in the nematode Caenorhabditis elegans. Genetics, 123(4), 755–769.
Sindhu, A. S., Maier, T. R., Mitchum, M. G., Hussey, R. S., Davis, E. L., & Baum, T. J. (2009). Effective and specific in planta RNAi in cyst nematodes: Expression interference of four parasitism genes reduces parasitic success. Journal of Experimental Botany, 60(1), 315–324. https://doi.org/10.1093/jxb/ern289.
Starr, J. L., Bridge, J., & Cook, R. (2002). Resistance to plant-parasitic nematodes: History, current use and future potential. In J. L. Starr, R. Cook, & J. Bridge (Eds.), Plant resistance to parasitic nematodes (1st ed., pp. 1–22). CABI. https://doi.org/10.1079/9780851994666.0001.
Szitenberg, A., Salazar-Jaramillo, L., Blok, V. C., Laetsch, D. R., Joseph, S., Williamson, V. M., Blaxter, M. L., & Lunt, D. H. (2017). Comparative genomics of apomictic root-knot nematodes: Hybridization, ploidy, and dynamic genome change. Genome Biology and Evolution, 9(10), 2844–2861. https://doi.org/10.1093/gbe/evx201.
Sukno, S. A., McCuiston, J., Wong, M. Y., Wang, X., Thon, M. R., Hussey, R., et al. (2007). Quantitative detection of double-stranded RNA-mediated gene silencing of parasitism genes in Heterodera glycines. Journal of nematology, 39(2), 145–152. /PMC/articles/PMC2586495/. Accessed 28 June 2021.
Triantaphyllou, A. C. (1973). Environmental sex differentiation of nematodes in relation to pest management. Annual Review of Phytopathology, 11(1), 441–462 www.annualreviews.org.
Trudgill, D. L., & Blok, V. C. (2001). Apomictic, polyphagous root-knot nematodes: Exceptionally successful and damaging biotrophic root pathogens. Annual Review of Phytopathology, 39(1), 53–77. https://doi.org/10.1146/annurev.phyto.39.1.53.
Villeneuve, A. M., & Meyer, B. J. (1987). Sdc-1: A link between sex determination and dosage compensation in C. elegans. Cell, 48(1), 25–37. https://doi.org/10.1016/0092-8674(87)90352-7.
Wang, S., & Kimble, J. (2001). The TRA-1 transcription factor binds TRA-2 to regulate sexual fates in Caenorhabditis elegans. EMBO Journal, 20(6), 1363–1372. https://doi.org/10.1093/emboj/20.6.1363.
Yadav, B. C., Veluthambi, K., & Subramaniam, K. (2006). Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Molecular and Biochemical Parasitology, 148(2), 219–222. https://doi.org/10.1016/j.molbiopara.2006.03.013.
Yonker, S. A., & Meyer, B. J. (2003). Recruitment of C. elegans dosage compensation proteins for gene-specific versus chromosome-wide repression. Development, 130(26), 6519–6532. https://doi.org/10.1242/dev.00886.
Zanetti, S., & Puoti, A. (2013). Sex determination in the Caenorhabditis elegans germline. In T. Schedl (Ed.), germ cell development in C. elegans (pp. 41–69). New York: Springer, New York, NY. https://doi.org/10.1007/978-1-4614-4015-4_3.
Zarkower, D. (2006). Somatic sex determination. WormBook. https://doi.org/10.1895/wormbook.1.84.1.
Acknowledgments
The authors are grateful to the Department of Entomology and Nematology, University of Florida for providing the opportunity to conduct this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
The article does not contain any human studies with human participants or animals performed by any authors.
Informed consent
Informed consent was obtained from all individual participants included in this study.
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baniya, A., Joseph, S., Duncan, L. et al. The role of Caenorhabditis elegans sex-determination homologs, Mi-sdc-1 and Mi-tra-1 in Meloidogyne incognita. Eur J Plant Pathol 161, 439–452 (2021). https://doi.org/10.1007/s10658-021-02335-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-021-02335-3