Abstract
Late blight caused by the oomycete pathogen Phytophthora infestans is one of the most destructive diseases in potato cultivation. To successfully colonize its host, P. infestans secretes a suite of effector proteins that undermine plant immunity, many of which contain a conserved N-terminal RXLR motif that strongly resembles the host targeting motif in effectors of the malaria parasite Plasmodium falciparum. In this study, we focus on three P. infestans clade 5 aspartic proteases (PiAPs) that are homologous to Plasmepsin V (PMV), a Pl. falciparum AP responsible for cleaving effectors prior to translocation into red blood cells. Malaria parasites expressing mutated PMV are impaired in effector translocation and are less virulent. To determine whether clade 5 PiAPs play similar roles in virulence, we characterized P. infestans transformants with either reduced or enhanced PiAP expression levels. Phytophthora infestans transformants with altered PiAP10 or PiAP12 expression were found to be impaired in mycelial growth and sporangia production, and are hampered in their virulence on potato leaves. This was not observed in PiAP11 transformants. Activity assays showed that PiAP10 and PiAP12 possess moderate protease activity, and can potentially cleave the RXLR effector PiAVR4, but not a PiAVR4 version with a mutated RXLR motif. These findings imply that P. infestans APs function in the proteolytic cleavage of RXLR effectors, and warrant further investigation to verify and confirm the role of clade 5 PiAPs in effector processing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant pathogenic microorganisms – including bacteria, fungi and oomycetes – secrete effector proteins that suppress host immune responses to facilitate successful host colonization. The potato late blight pathogen Phytophthora infestans – as well as related oomycete species – harbor an extended arsenal of effector proteins, of which the majority contains the conserved N-terminal RXLR (Arginine-x-Leucine-Arginine) peptide motif (Anderson et al. 2015). These so-called RXLR effectors are considered to be translocated into host cells via specialized infection structures, such as haustoria. Multiple studies show that the RXLR motif is essential for the delivery of P. infestans effectors into host cells, and that mutation of the RXLR motif hampers effector translocation (Whisson et al. 2007; Schornack et al. 2009). It was only recently shown that RXLR effectors indeed shuttle into host cells by visualizing an mRFP-tagged RXLR effector that accumulates in the plant cell nucleus (Wang et al. 2017). The exact mechanism that supports translocation of RXLR effectors, however, has not been fully resolved.
The Phytophthora RXLR motif strongly resembles the host translocation motif RXLXE/Q/D of PEXEL effectors secreted by the malaria pathogen Plasmodium falciparum. Studies by Boddey et al. (2010) and Russo et al. (2010) showed that the N-terminal RXLXE/Q/D motif of Plasmodium PEXEL effectors is cleaved after the Leucine (L) residue by the aspartic protease Plasmepsin V (PMV) prior to host translocation into red blood cells. PMV harbors a C-terminal transmembrane domain, and was found to reside in the endoplasmic reticulum (ER) of the pathogen (Russo et al. 2010). Inhibition of PMV activity by using either HIV inhibitors, the aspartic protease inhibitor pepstatin or a PEXEL-mimetic inhibitor, was shown to impact effector export and to result in loss of virulence (Russo et al. 2010; Sleebs et al. 2014). Translocation of cleaved PEXEL effectors into host cells is mediated by the Plasmodium translocon of exported proteins (PTEX) complex, which comprises as core components the exported protein EXP2, the AAA+-ATPase HSP101 and PTEX150, a novel Plasmodium protein with unknown function (Elsworth et al. 2014).
The strong resemblance of host translocation motifs (RXLXE/Q/D versus RXLR) in effectors of Phytophthora spp. and Pl. falciparum suggested that both organisms make use of similar export mechanisms to deliver effectors into host cells (Bhattacharjee et al. 2006; Bouwmeester et al. 2011). The current predominant model emphasizes cleavage of the RXLR motif to initiate effector translocation into host cells. (Bouwmeester et al. 2011; Petre and Kamoun 2014; Wang et al. 2017; Wawra et al. 2017). Phytophthora genomes, however, lack genes encoding homologs of PTEX components and as such the mechanism by which RXLR effectors are translocated remains elusive (Bouwmeester et al. 2011). Another model suggests that the RXLR motif binds to phosphatidylinositol 3-phosphate (PI(3)P) in host membranes, thereby activating effector translocation by pathogen-independent lipid raft-mediated endocytosis (Kale et al. 2010).
Kay et al. (2011) identified 12 aspartic proteases (APs) in P. infestans, including three clade 5 PiAPs – i.e. PiAP10, PiAP11 and PiAP12 – that are homologous to Pl. falciparum PMV. APs are endopeptidases usually containing two active aspartate residues in their catalytic site that bind a water molecule to initiate cleavage of protein substrates. Well-known eukaryotic APs are pepsins, cathepsins and renins (Revuelta et al. 2014). Several plant APs have been shown to play keys role in pathogen defense and abiotic stress response (Simões and Faro 2004). In fungi, APs are involved in various physiological processes, including fitness and growth. Inhibition of aspartic proteases in diverse plant pathogenic fungi, such as Alternaria and Fusarium species, was shown to result in growth inhibition (Dash et al. 2001). Examples of APs involved in pathogen-host interactions are HIV-1, which functions in protein cleavage of structural components to generate infectious HIV virions, and the secretory aspartic proteases SAP1–10 of the yeast Candida albicans that are involved in pathogenicity and cleavage of host proteins (Craig et al. 1998; Monod and Borg-von Zepelin 2002; Naglik et al. 2003; Silva et al. 2014). ASP5, an AP from the zoonotic parasite Toxoplasma gondii, was found to be located in the Golgi apparatus and plays a key role in the cleavage of GRA effectors at a PEXEL-like motif. Deletion of ASP5 resulted in reduced parasite fitness and virulence (Hammoudi et al. 2015; Cassone et al. 2016).
This study focuses on P. infestans clade 5 aspartic proteases (PiAPs) that are homologues to Plasmodium PMV. Here, we determined the involvement of three PiAPs in virulence by functionally characterizing P. infestans transformants with altered PiAP gene expression levels. In order to test whether the clade 5 PiAPs have a similar role in effector modification as PMV, we assessed the potential activity of the PiAPs in effector cleavage using the P. infestans RXLR effector AVR4 as substrate.
Materials and methods
Phytophthora culture conditions and plant infection assays
Phytophthora infestans isolates were grown at 18 °C on rye sucrose (RS) agar plates supplemented with appropriate antibiotics. P. infestans transformants were selected and grown on RS agar plates supplemented with 5 mg/L geneticin (G418). Zoospores were isolated by flooding two-week old mycelium with ice-cold water for 2–3 h. Inoculum concentrations were adjusted to 1 × 105 zoospores/ml. Five-week old potato plants (cv Désirée, [R0]) were spray-inoculated with P. infestans zoospore suspensions until run-off. Inoculation assays on detached Nicotiana benthamiana and potato leaves were performed by placing 10 μl droplets of inoculum on abaxial leaf sides. Mock-inoculation was performed with water. Inoculated plants were incubated at 18 °C and high humidity. Lesion sizes were determined at 5 days post inoculation (dpi) as described by Vleeshouwers et al. (1999). Statistical analysis was performed using One-way ANOVA with Tukey’s HSD post-hoc tests (p < 0.05).
Nucleic acid isolation and quantitative RT-PCR
Phytophthora infestans DNA was extracted using TRIzol reagent, and RNA was isolated with a Quick-RNA Mini-prep kit according to the manufacturer’s instructions. cDNA was synthesized from 1 μg of total RNA using oligo(dT) primers and M-MLV reverse transcriptase. Quantitative RT-PCR was performed in triplicate on an ABI7300 SDS real-time qPCR machine. Transcript levels of PiAPs were quantified using gene-specific primers and PiActA as endogenous control (Table S1). Data were analyzed based on the 2-ΔΔCT method (Schmittgen and Livak 2008). Samples were taken at eight time points from 0 to 72 h post inoculation (hpi).
Plasmid construction
Full-length sequences of PiAP10, −11 and −12 were PCR-amplified from genomic DNA of P. infestans isolate 88069 using proof-reading DNA polymerase and primers that generate restriction sites (Table S1). PCR fragments were introduced by directional cloning into plasmid pMCherryN, which carries a geneticin resistance cassette and a mCherry sequence under control of a ham34 promoter (Ah-Fong and Judelson 2011). Sequences encoding PiAP domains (PiAP1066–413, PiAP1196–441, PiAP1291–430) were amplified from cDNA with primers containing a 6His sequence (Table S1). Sequences encoding C-terminal 6His/3Myc-tagged PiAVR4RFLR (AA25–287) and the RXLR mutant version PiAVR4AAAA (RFLR→AAAA) were obtained via gene synthesis. Gene fragments were introduced by restriction digestion to plasmid pFLAG-ATS. Plasmid PMV-3His encoding Pl. falciparum Plasmepsin V was provided by Boddey et al. (2010). All constructs were introduced to E. coli strain BL-21 for protein production. Full length PiAP sequences amplified from cDNA were introduced in plasmid pENTR/D-TOPO, and subsequently in the binary plasmid pGWB5 via LR reaction (Invitrogen). The recombinant plasmids were transformed by electroporation into A. tumefaciens strain AGL1 for Agrobacterium–mediated transient transformation assays.
Transformation of Phytophthora infestans
Protoplasts of P. infestans T35–3 were obtained by treating 2-days-old mycelium for 1 h with 5 mg/ml cellulase and 10 mg/ml β-glucanase. Prior to transformation, 25 μg plasmid DNA was mixed with 60 μl lipofectin and incubated for 15 min at RT. Protoplasts were transformed using the PEG transformation protocol as described by Ah-Fong et al. (2008). Transformed protoplasts were kept 24 h in liquid RS medium containing 5 mg/L geneticin (G418) and subsequently plated on RS agar plates with geneticin. Colonies were cultured on fresh RS agar plates and covered with a thin layer of RS agar supplemented with 5 mg/L geneticin. Emerging colonies were isolated and grown for 2 weeks on selective medium. Mycelial growth and sporangia production were determined 12 days after inoculation of a mycelial plug (5 mm in diameter) in the centre of a fresh RS agar plate.
Agroinfiltration assays
Agrobacterium tumefaciens AGL1 strains carrying binary vectors were grown overnight at 37 °C in LB medium supplemented with appropriate antibiotics. Bacterial pellets were resuspended to an OD600 of 1.0 in induction medium (per L: 20 g sucrose, 5 g MS basal salt mixture, 1.95 g MES, 200 mM acetosyringone, pH 5.6). Agroinfiltrations were performed on abaxial sides of N. benthamiana leaves using a needleless syringe. Plants were kept in a climate chamber at 21 °C and a 70% relative humidity.
Electrolyte leakage assays
Six leaf disks (9 mm in diameter) were excised per agroinfiltrated area and incubated for 3 h in 5 ml MQ under gentle agitation. After incubation, sample conductivity was measured using a digital conductivity meter equipped with LabX direct PH 2.1 software (Mettler Toledo). Total conductivity was measured after boiling leaf disks in solution for 15 min. Electrolyte leakage was calculated by dividing sample conductivity by the total conductivity. Experiments were repeated in triplicate each consisting three replicates per sample.
Protein production and purification
Recombinant E. coli strains were grown overnight as pre-cultures in 25 ml LB medium containing appropriate antibiotics at 37 °C. Individual pre-cultures were used to inoculate in 1 L of LB medium, and grown at 37 °C for 2 h to an OD600 of 0.5. Recombinant protein production and purification was performed as described in Fig. S1. Protein production was induced by adding 0.35 mM IPTG and cultures were subsequently grown for 4 h at 30 °C. Cells were collected by centrifugation at 5000 rpm for 30 min, and lysed in 40 ml lysis buffer (per 100 ml: 7.4 g sodium phosphate, 10 ml glycerol, 300 ml lysozyme, 6.26 ng DNAse) for 1 h at room temperature (RT, ~21 °C). Samples were snap-frozen in liquid nitrogen and thawed at RT. Supernatants (soluble protein fractions) were obtained by centrifugation at 14000 rpm for 45 min. Pellets were resuspended in 5 ml denaturing buffer (100 mM NaH2PO4, 100 mM Tris-HCl, 6 M GdnHCl, pH 8.0), incubated for 40 min at RT, and subsequently centrifuged at 14000 rpm for 45 min (denatured protein fractions). Protein fractions were stored at −20 °C. Protein purification was performed with Ni-NTA agarose beads. Protein extracts were mixed with beads (200 μl beads per 1 ml extract) and incubated for 2 h at 4 °C. Beads were collected by centrifugation at 800 rpm for 5 min and rinsed twice with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0). Bound proteins were eluted by incubation for 15 min at 4 °C with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). Purified proteins from soluble fractions were dialysed (Spectra/Por 3 RC dialysis tubing, MWCO 3,5 kDa, Spectrum Labs, USA) against 20 volumes of dialysis buffer (150 mM NaCl, 50 mM Tris-Cl, pH 8.0) overnight at 4 °C. Denatured protein fractions were washed and eluted in buffers additionally supplemented with 6 M guanidine hydrochlorite (GdnHCl). Purified denatured proteins were step-wise refolded using dialysis buffer with decreasing GdnHCl concentrations (6, 5, 4, 3, 2, 1, 0.5 and 0 M). Dialysis buffer was changed every 12–15 h. Purified proteins were stored in 20% glycerol at −80 °C.
Protein detection
Proteins samples were boiled for 5 min in SDS-loading buffer and separated on 4–20% gradient Mini-PROTEAN TGX Stain-free Precast protein gels (Bio-Rad). After electrophoresis, proteins were transferred to Immuno-Blot polyvinylidene difluoride membranes (Bio-Rad). Proteins were detected by incubating membranes with α-His or α-FLAG antibodies (diluted to 1:5000 in TBST) in 5% skimmed milk for 1 h at RT. Western blot signals were visualized using SuperSignal West Dura substrate (Thermo Fisher Scientific) using a ChemiDoc XRS system. Protein blots were subsequently stained with Coomassie Brilliant Blue R250.
Zymography and in vitro cleavage assays
In-gel zymography assays were performed according to Hassani et al. (2014) with slight modifications. In brief, 12% polyacrylamide gels containing 1.5 mg/ml gelatin were loaded with purified soluble and refolded PiAP proteins at a concentration of 10 μg. Trypsin was used as a positive control for protein digestion. Trypsin activity was inhibited using 50 μΜ protease inhibitor mix (cOmplete ULTRA, Sigma-Aldrich), and AP activity using 5 μΜ pepstatin A (Sigma-Aldrich). After electrophoresis, gels were incubated with 1% Triton X-100 for 1 h at RT, and subsequently washed overnight in 50 mM Tris-Cl pH 7.4, 5 mM, CaCl2 and 1 mM ZnCl, at RT under constant agitation. Gels were stained with Coomassie Brilliant Blue R250.
In vitro activity assays were performed according to Boddey et al. (2010), with modifications. Purified PiAPs and PMV (2 μg) were mixed with soluble PiAVR4RFLR or PiAVR4AAAA (1 μg) in 100 μl activity buffer (25 mM Tris, 25 mM MES, pH 6.5), and incubated for 1 h at RT under constant shaking. Hereafter, samples were boiled for 5 min in SDS-loading buffer prior to SDS-PAGE. Proteases and substrate proteins were detected with immunoblotting using α-FLAG and α-His antibodies, respectively.
Multiple sequence alignment and phylogenetic analysis
Protein sequences of P. infestans APs and Pl. falciparum Plasmepsin V were retrieved from Kay et al. 2011, and aligned with Clustal Omega or MAFFT using default settings. A phylogenetic tree was constructed with MEGA6 using maximum likelihood analysis. Tree robustness was assessed by bootstrap analysis using 1000 replicates.
Results
PiAP expression is enhanced in pre-infection stages and peaks during host colonization
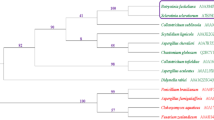
To gain insight into the biological functions of clade 5 PiAPs (Fig. 1a), we first assessed the expression of PiAP10, PiAP11, and PiAP12 in four different P. infestans life stages, i.e. mycelium, sporangia, zoospores and germinating cysts. Quantitative RT-PCR using gene-specific primers revealed that clade 5 PiAPs are expressed in all developmental stages, with highest transcript levels in germinating cysts (Fig. 1b). Expression of PiAP10 and PiAP11 was also found to be induced in zoospores, whereas PiAP12 transcription is elevated in sporangia. Next, we determined PiAP expression during potato leaf infection, i.e. at time points 0 to 72 hpi. The three PiAPs were found to be expressed during all time points, and show remarkably similar profiles including a sharp increase in expression at 36 hpi (Fig. 1c). This distinct peak in expression could be related to full establishment of disease or to haustoria development (Zuluaga et al. 2016).
Phylogeny of aspartic proteases in Phytophthora infestans and expression of clade 5 PiAPs in different P. infestans life stages and during potato infection. a: Phylogenetic tree of P. infestans APs based on full-length amino acid sequences. Clade 5 PiAPs clustering with Plasmodium falciparum Plasmepsin V are outlined in blue. Numbers at nodes indicate bootstrap values representing the level of clade support inferred by Maximum Likelihood. b: Expression profiles of PiAP10, −11 and −12 in different life stages, i.e. mycelium (MY), sporangia (SP), zoospores (ZO), and germinating cysts (GC). Expression levels were quantified by qRT-PCR relative to the expression in mycelium (set at 1) and with PiActA as endogenous control. c: Expression profiles of PiAP10, −11 and −12 during infection of potato. Leaves of spray-inoculated plants were harvested at different time points between 0 and 72 h post-inoculation. RNA was isolated and expression levels were quantified by qRT-PCR relative to the expression at time point 0 (set at 1) . Bars represent averages ± SD of two biological replicates
Altered PiAP expression in P. infestans affects colony growth
To further investigate the role of clade 5 PiAPs, we transformed P. infestans isolate T35–3 with constructs encoding full length PiAPs harboring mCherry tags. Transformants selected for their resistance to geneticin were verified by PCR, and subsequently analyzed by qRT-PCR to determine PiAP expression levels. Transformants with a > 25% higher level in PiAP expression were considered to be overexpressing lines (PiAP-OX), whereas those with a reduction in gene expression of at least 25% were selected as silenced lines (PiAP-Sil). In total, we obtained 16 PiAP-OX lines; i.e. eight lines for PiAP10, four for PiAP11 and four for PiAP12. Transformation also resulted in seven PiAP-Sil lines; three silenced lines for PiAP10, two for PiAP11 and two for PiAP12 (Fig. 2). In none of the Sil or OX lines transformation of the target gene affected the expression of the other two clade 5 PiAPs. Two P. infestans transformants that only express mCherry were selected as empty vector controls (mCh22 and mCh25). Phenotypic characterization showed that transformants enhanced or silenced in PiAP10 and PiAP12 expression are affected in colony morphology. All PiAP10- and PiAP12-Sil lines showed reduction in mycelial growth and sporangia production (Fig. 3), whereas such a clear effect on development was not found among PiAP10- and PiAP12-OX lines. In contrast, both the PiAP11-Sil and -OX lines seem to be slightly enhanced in growth and production of sporangia (Fig. 3), but no correlation could be found between the degree of silencing or overexpression and the observed phenotypic effects. All PiAP11 transformants, regardless of their transgene expression level, were able to produce zoospores in amounts comparable to that of the control lines. Taken together, these results suggest that altered expression levels of PiAP10 and PiAP12 affects P. infestans growth and sporangia production, while altered expression of PiAP11 had no obvious effects.
PiAP-overexpressing and -silenced transformants. PiAP10, −11, and −12 expression in P. infestans transformants was analysed by qRT-PCR and transcript levels were quantified relative to the level in the recipient strain T35–3 (R) (set at 1). PiActA was used as endogenous control. Transformants with increased or decreased PiAP transcript levels are referred to as overexpressing (-OX) or silenced (-Sil) lines, respectively. Bars represent averages ± SD of two biological replicates
Altered PiAP expression affects P. infestans growth and sporulation. a: Colony size and b: sporangia production of P. infestans -OX and -Sil lines at 9 dpi. Lines transformed with the empty vector pMCherryN (mCh22 and mCh25) were included as controls. Bars represent averages ± SD of two biological repeats. Different letters indicate significant differences (One-way ANOVA, p < 0,05)
P. infestans transformants with altered PiAP10 or PiAP12 expression are hampered in virulence
In a next step, we assayed whether alteration of PiAP expression influences P. infestans virulence. Potato leaves were inoculated with similar amounts of zoospores, and infection efficiency and lesion formation were measured after 5 days. Control lines mCh22 and mCh25 were found to behave in a similar way as the recipient isolate T35–3. In contrast, potato leaves inoculated with P. infestans lines that overexpress either PiAP10 or PiAP12, resulted in significantly smaller lesions (Fig. 4a). It should be noted that these OX-lines also have lower infection efficiencies. This is largely due to the fact that inoculation with these lines frequently resulted in cell death (Fig. 4b). Transformants overexpressing PiAP11 (PiAP11-OX) developed lesions comparable to the control, with no reduction in efficiency and no induction of cell death (Fig. 4). PiAP10- and PiAP12-Sil lines have a low infection efficiency and show significantly reduced lesion sizes on inoculated potato leaves, but did not induce cell death (Fig. 4). Similar to the PiAP11-OX lines, transformants silenced for PiAP11 did not show any reduction in infection rate or lesion size in comparison to the control lines (Fig. 4).
Infection assay of P. infestans PiAP transformants on detached potato leaves. A: Lesion sizes on potato leaves at 5 dpi. Data are an average of three biological repeats ± SD (n = 18). Different letters indicate significant differences (One-way ANOVA, p < 0,05). B: Infection efficiency of PiAP transformants. Green and white sectors in the pie charts depict the percentages of successful and unsuccessful infection, respectively. Numbers connected to the white sectors represent percentages of cell death observed among unsuccessful infections at 5 dpi. Data are an average of three biological repeats (n = 30)
Expression of PiAP10 and PiAP12 in Nicotiana benthamiana impairs colonization by P. infestans
To investigate if the observed cell death induced by PiAP10- and PiAP12-OX lines is pathogen independent, we transiently expressed PiAPs in N. benthamiana leaves by agro-transformation. No cell death was observed by the naked eye in PiAP-expressing zones at 5 dpi. Empty vector (EV) and the cell death-inducing protein CRN2 (Torto et al. 2003) were used as negative and positive controls, respectively. In addition, we monitored ion leakage as a measure of cell death. Agroinfiltrated leaves that transiently express PiAP10 and PiAP12 showed an increase in ion leakage compared to the negative control, but this was not comparable to the strong response initiated by the positive control (Fig. 5a). Transient expression of PiAP11 had only a minor effect. Next, we performed infection assays on agroinfiltrated leaves with P. infestans zoospores. In planta expression of PiAP10 and PiAP12 significantly reduced leaf colonization, while leaves that express PiAP11 were unaffected in lesion growth (Fig. 5b-c).
PiAP10 and PiAP12 inhibit growth of Phytophthora infestans in Nicotiana benthamiana.A: Ion leakage in N. benthamiana leaves transiently expressing PiAPs compared to empty vector (EV) and CRN2 control. Conductivity values were measured 5 days after agroinfiltration. Data are an average of three biological repeats ± SD (n = 15). Different letters indicate significant differences with the control (One-way ANOVA, p < 0,05). B: Mean sizes of P. infestans lesions at 5 dpi on N. benthamiana leaves transiently expressing PiAPs. Data are averages of three biological repeats ± SD (n = 30). Different letters indicate significant differences with the control (One-way ANOVA, p < 0,05). C: Images taken under UV light showing leaves of N. benthamiana that were agroinfiltrated with the PiAP constructs (as in B) and subsequently inoculated with P. infestans
PiAP10 and PiAP12 potentially cleave P. infestans RXLR effector AVR4
To assess whether clade 5 PiAPs function in RXLR effector cleavage, we performed in vitro experiments with heterologously produced aspartic domains of PiAP10, PiAP11 and PiAP12. FLAG/His-tagged gene fragments encoding aspartic domains (i.e. PiAP10; AA66–413, PiAP11; AA96–441, PiAP12; AA91–430) were constructed and expressed in E. coli (Fig. 6a). Recombinantly produced and purified PiAPs were first analyzed for proteolytic activity by so-called in-gel zymography; assays based on enzymatic degradation of high-molecular-weight substrates integrated in polyacrylamide gels that can be visualized by electrophoresis (Choi et al. 2009). In our assays we used gelatin, a mixture of proteins derived after enzymatic hydrolysis of collagen. As control we included trypsin, a serine protease that cleaves between lysine (K) and arginine (R) residues. Results show that trypsin was able to digest gelatin efficiently, as indicated by the strong discoloration of the gel (Fig. S2). Discoloration of the gel was also observed upon incubation with both soluble and refolded PiAP10, −11, and −12, although much weaker as observed with trypsin (Fig. S2). When we added the aspartic protease inhibitor pepstatin no discoloration was observed, suggesting that clade 5 PiAPs have AP activity.
PiAP10 and PiAP12 can cleave P. infestans effector AVR4. a: Cartoons of recombinantly produced PiAPs, PMV and PiAVR4. The red line in AVR4RFLR and AVR4AAAA indicates the position of the RXLR motif. Boxes outlined in blue indicate the tags (M: Myc, F: FLAG, H: His). b: AVR4RFLR is cleaved by PiAP10 and PiAP12, but not by PiAP11 and PMV. Arrowheads point to the PiAVR4 fragments obtained after cleavage. c: PiAP10 and PiAP12 are unable to cleave the mutated version AVR4AAAA. d: Soluble and refolded PiAPs produced in E. coli and detected on immunoblots incubated with FLAG antibodies. Protein sizes in kDa are indicated
Based on the fact that the aspartic protease PMV of the malaria parasite Pl. falciparum cleaves PEXEL effectors after the L residue within the motif RXLXE/Q/D, we performed activity assays to determine if PiAPs can cleave RXLR effectors in a similar fashion. For this purpose, we heterologously produced a tagged version of the P. infestans RXLR effector AVR4 (AVR4RFLR). Two bands were detected by immunoblotting, one with an expected size of approximately 40 kDa and a lower band at 25 kDa that could be a degradation product. Incubation of AVR4RFLR with PiAP10 and PiAP12 resulted in the appearance of a third band slightly smaller than AVR4RFLR. The size of this band is around 35 kDa, which is the expected size of AVR4 when cleaved in or near the RXLR motif. A similar sized band was not detected in assays using PiAP11 and PMV, suggesting some sort of enzyme specificity (Fig. 6b). These results suggest enzymatic cleavage of AVR4 at the N-terminus.
To further validate the involvement of the RXLR motif in effector cleavage we produced a version of AVR4 in which the RXLR motif is mutated into AAAA (AVR4AAAA), which migrated as a single 40 kDa band. Activity assays using AVR4AAAA as substrate did not result in a comparable sized extra band of 35 kDa upon incubation with any of the recombinant PiAPs (Fig. 6c). These results imply that AVR4 is cleaved in the RXLR motif, or that the RXLR motif is important for the cleavage process.
Discussion
APs are an important class of enzymes involved in virulence of several mammalian parasites, and shown to play key roles in host cell manipulation to facilitate disease. Well-studied examples are PMV from Pl. falciparum that modifies PEXEL effectors to activate translocation into the host erythrocytes and ASP5 from T. gondii, which modifies effectors at the PEXEL-like motif and plays a role in the pathogen fitness and virulence (Hammoudi et al. 2015; Boddey et al. 2010; Russo et al. 2010). A large amount of effectors secreted by plant pathogenic oomycetes harbour a N-terminal RXLR motif sequence highly similar in sequence to the PEXEL motif. This led to the tempting hypothesis that a similar mechanism exists for RXLR effector translocation. Early studies showed that PEXEL and RXLR motifs are interchangeable, however, PMV was not able to cleave RXLR effectors (Bhattacharjee et al. 2006). In a later study Bhattacharjee et al. (2012a) showed that in Plasmodium, export of PEXEL effectors is facilitated by passage via the ER. Their findings lead to a model in which host targeting of PEXEL effectors is mediated by binding to PI(3)P in the parasite’s ER and is independent of PMV-mediated PEXEL cleavage (Bhattacharjee et al. 2012a, b). A recent study by Boddey et al. (2016) could not find sufficient evidence to support this model and showed that the modification of the PEXEL effectors by PMV is the essential step to initiate effector translocation.
RXLR effectors are found to accumulate around haustoria, the site where they supposedly enter the host cells. Several studies have been focussing on the mechanism responsible for effector translocation into the host cell, and most of them indicated that the RXLR motif is essential (Schornack et al. 2009). However, several studies show that the presence of the pathogen is not necessary for the export of RXLR effectors. The P. sojae effector AVR1 was found to enter host cells via a mechanism independent of the pathogen (Dou et al. 2008). However, experiments showing this autonomous uptake were not repeatable (Wawra et al. 2013). A different mechanism of translocation that is mediated by external PI(3)P binding of the RXLR motif of effectors has been suggested (Kale et al. 2010). This model of effector translocation has also been under heavy debate, with more recent studies indicating that the RXLR motif is not sufficient for PI(3)P binding and that the uptake is pathogen-dependent (Yaeno and Shirasu 2013; Petre and Kamoun 2014; Petre et al. 2016).
In this study we focused on three clade 5 APs of P. infestans that show similarity to PMV of Pl. falciparum. PiAPs and PfPMV share structural features, including signal peptide and propeptide regions. Like PMV, PiAPs harbor a C-terminal transmembrane domain, which in PfPMV confers ER localization (Fig. S3) (Russo et al. 2010; Tarr and Osborne 2015).
We hypothesized that these APs may be involved in virulence of P. infestans by modifying RXLR effectors. As a first approach, the potential role of PiAPs on P. infestans virulence was examined by altering their expression. Overexpression of PiAPs did not have a severe effect on growth and sporulation. However, silencing of PiAP10 and PiAP12 resulted in reduced colony growth and sporulation suggesting that these APs have a role in pathogen fitness. Similar results have been reported in other studies, where inhibition of APs resulted in growth inhibition of several fungal species (Dash et al. 2001), while deletion of ASP5 in T. gondii led to reduced fitness of the pathogen (Hammoudi et al. 2015).
To study the role of the PiAPs in virulence, we performed disease assays with transformants with silenced PiAP expression. Lines that were silenced in PiAP10 or PiAP12 – but not PiAP11 – expression were found to be reduced in virulence, as indicated by the formation of significantly smaller lesions on potato leaves. Inoculation of potato leaves with transformants overexpressing PiAP10 or PiAP12 induced cell death and resulted in smaller lesions. Similar results were not observed when using PiAP11-overexpressing lines. Our agroinfiltration assays showed that transient expression of PiAP10 and PiAP12 in N. benthamiana leaves reduces lesion formation. This could suggest that PiAPs can be active inside host cells, somehow stimulating plant defense hindering host colonization. Protein structure prediction suggests that PiAP10 and PiAP12 are membrane-bound and may function as a homodimers. PiAP11 is as well membrane-bound, but it lacks the essential cysteine residues to form dimers (Kay et al. 2011). We hypothesize that this could be a potential explanation for the observed differences between PiAP10 and −12, and PiAP11.
To determine whether clade 5 PiAPs have a similar biochemical function as PMV, we performed in vitro experiments with heterologously produced aspartic domains of PiAP10, PiAP11 and PiAP12. Enzymatic activity assays using gelatin as a general substrate showed that the aspartic domains of all three clade 5 PiAPs harbour proteolytic activity and this was verified by inhibition of their activity with the general AP inhibitor pepstatin. Activity assays using recombinant PiAPs suggest that both PiAP10 and PiAP11 are capable to cleave the P. infestans RXLR effector AVR4. Furthermore, we show that the RXLR motif in AVR4 is important, since mutation of this motif resulted in loss of enzymatic cleavage. The ability of clade 5 PiAPs to cleave at neutral pH is potentially due to substitution of an alanine residue, which in human renin contributes to enzyme activity over an extended pH range (Yamauchi et al. 1988; Fig. S3).
Recently, Wawra et al. (2017) showed that the RXLR effector AVR3 is cleaved in the N-terminus before secretion. In that study, however, none of the clade 5 PiAPs was found to modify the effector and this is in contrast to our findings. This could be due to differences in experimental set-up of the activity assays or to the fact that a different RXLR effector is used as substrate. It is very well possible that not all RXLR effectors are modified by the same enzyme.
Taken together, our results indicate that PiAP10 and PiAP12 play a role in P. infestans virulence and that they are key components in the infection process. Based on these findings, we propose a model in which specific PiAPs act as key components in the modification of one or a particular group of RXLR effectors and therefore promote disease in a very precise and balanced manner and that alteration of this balance affects the fitness and the virulence of the pathogen. Reduced levels of PiAP interferes with effector secretion which results in reduced virulence. Overproduction leads to uncontrolled activity and causes imbalanced effector export, which induces cell death. To unravel the underlying mechanism, the role of PiAPs in RXLR effector processing needs to be confirmed by further optimizing the activity assays and by using a wide diversity of RXLR effectors as substrate.
References
Ah-Fong, A. M. V., & Judelson, H. S. (2011). Vectors for fluorescent protein tagging in Phytophthora: tools for functional genomics and cell biology. Fungal Biology, 115, 882–890.
Ah-Fong, A. M. V., Bormann-Chung, C. A., & Judelson, H. S. (2008). Optimization of transgene-mediated silencing in Phytophthora infestans and its association with small-interfering RNAs. Fungal Genetics and Biology, 45, 1197–1205.
Anderson, R. G., Deb, D., Fedkenheuer, K., & McDowell, J. M. (2015). Recent progress in RXLR effector research. Molecular Plant Microbe Interactions, 28, 1063–1072.
Bedi, R. K., Patel, C., Mishra, V., Xiao, H., Yada, R. Y., & Bhaumik, P. (2016). Understanding the structural basis of substrate recognition by Plasmodium falciparum plasmepsin V to aid in the design of potent inhibitors. Scientific Reports, 6, 31420.
Bhattacharjee, S., Hiller, N. L., Liolios, K., Win, J., Kanneganti, T.-D., Young, C., Kamoun, S., & Haldar, K. (2006). The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathogens, 2, e50.
Bhattacharjee, S., Stahelin, R. V., Speicher, K. D., Speicher, D. W., & Haldar, K. (2012a). Endoplasmic reticulum PI(3)P lipid binding targets malaria proteins to the host cell. Cell, 148, 201–212.
Bhattacharjee, S., Stahelin, R. V., & Haldar, K. (2012b). Host targeting of virulence determinants and phosphoinositides in blood stage malaria parasites. Trends in Parasitology, 28, 555–562.
Boddey, J. A., Hodder, A. N., Gunther, S., Gilson, P. R., Patsiouras, H., Kapp, E. A., Pearce, J. A., de Koning-Ward, T. F., Simpson, R. J., Crabb, B. S., & Cowman, A. F. (2010). An aspartyl protease directs malaria effector proteins to the host cell. Nature, 463, 627–631.
Boddey, J. A., O'Neill, M. T., Lopaticki, S., Carvalho, T. G., Hodder, A. N., Nebl, T., Wawra, S., van West, P., Ebrahimzadeh, Z., Richard, D., Flemming, S., Spielmann, T., Przyborski, J., Babon, J. J., & Cowman, A. F. (2016). Export of malaria proteins requires co-translational processing of the PEXEL motif independent of phosphatidylinositol-3-phosphate binding. Nature Communications, 7, 10470.
Bouwmeester, K., Meijer, H. J. G., & Govers, F. (2011). At the frontier; effectors crossing the Phytophthora-host interface. Frontiers in Plant Science, 2, 75.
Cassone, A., Vecchiarelli, A., & Hube, B. (2016). Aspartyl proteinases of eukaryotic microbial pathogens: From eating to heating. PLoS Pathogens, 12, e1005992.
Choi, N.-S., Kim, B.-H., Park, C.-S., Han, Y. J., Lee, H. W., Choi, J. H., Lee, S.-G., & Song, J. J. (2009). Multiple-layer substrate zymography for detection of several enzymes in a single sodium dodecyl sulfate gel. Analytical Biochemistry, 386, 121–122.
Craig, C., Race, E., Sheldon, J., Whittaker, L., Gilbert, S., Moffatt, A., Rose, J., Dissanayeke, S., Chirn, G. W., Duncan, I. B., & Cammack, N. (1998). HIV protease genotype and viral sensitivity to HIV protease inhibitors following saquinavir therapy. AIDS, 12, 1611–1618.
Dash, C., Ahmad, A., Nath, D., & Rao, M. (2001). Novel bifunctional inhibitor of xylanase and aspartic protease: Implications for inhibition of fungal growth. Antimicrobial Agents and Chemotherapy, 45, 2008–2017.
Dou, D., Kale, S. D., Wang, X., Jiang, R. H. Y., Bruce, N. A., Arredondo, F. D., Zhang, X., & Tyler, B. M. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. The Plant Cell, 20, 1930–1947.
Elsworth, B., Matthews, K., Nie, C. Q., Kalanon, M., Charnaud, S. C., Sanders, P. R., Chisholm, S. A., Counihan, N. A., Shaw, P. J., Pino, P., Chan, J.-A., Azevedo, M. F., Rogerson, S. J., Beeson, J. G., Crabb, B. S., Gilson, P. R., & de Koning-Ward, T. F. (2014). PTEX is an essential nexus for protein export in malaria parasites. Nature, 511, 587–591.
Hammoudi, P.-M., Jacot, D., Mueller, C., Di Cristina, M., Dogga, S. K., Marq, J.-B., Romano, J., Tosetti, N., Dubrot, J., Emre, Y., Lunghi, M., Coppens, I., Yamamoto, M., Sojka, D., Pino, P., & Soldati-Favre, D. (2015). Fundamental roles of the Golgi-associated Toxoplasma aspartyl protease, ASP5, at the host-parasite interface. PLoS Pathogens, 11, e1005211.
Hassani, K., Shio, M. T., Martel, C., Faubert, D., Olivier, M., & Langsley, G. (2014). Absence of metalloprotease GP63 alters the protein content of Leishmania exosomes. PLoS One, 9, e95007.
Kale, S. D., Gu, B., Capelluto, D. G. S., Dou, D., Feldman, E., Rumore, A., Arredondo, F. D., Hanlon, R., Fudal, I., Rouxel, T., Lawrence, C. B., Shan, W., & Tyler, B. M. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell, 142, 284–295.
Kay, J., Meijer, H. J. G., ten Have, A., & van Kan, J. A. L. (2011). The aspartic proteinase family of three Phytophthora species. BMC Genomics, 12, 254.
Monod, M., & Borg-von Zepelin, M. (2002). Secreted aspartic proteases as virulence factors of Candida species. Biological Chemistry, 383, 1087–1093.
Naglik, J. R., Challacombe, S. J., & Hube, B. (2003). Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiology and Molecular Biology Reviews, 67, 400–428.
Petre, B., & Kamoun, S. (2014). How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biology, 12, e1001801.
Petre, B., Kopischke, M., Evrard, A., Robatzek, S., & Kamoun, S. (2016). Cell re-entry assays do not support models of pathogen-independent translocation of AvrM and AVR3a effectors into plant cells. bioRxiv. https://doi.org/10.1101/038232.
Revuelta, M. V., van Kan, J. A. L., Kay, J., & ten Have, A. (2014). Extensive expansion of A1 family aspartic proteinases in fungi revealed by evolutionary analyses of 107 complete eukaryotic proteomes. Genome Biology and Evolution, 6, 1480–1494.
Russo, I., Babbitt, S., Muralidharan, V., Butler, T., Oksman, A., & Goldberg, D. E. (2010). Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature, 463, 632–636.
Schmittgen, T. D., & Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nature Protocols, 3, 1101–1108.
Schornack, S., Huitema, E., Cano, L. M., Bozkurt, T. O., Oliva, R., Van Damme, M., Schwizer, S., Raffaele, S., Chaparro-Garcia, A., Farrer, R., Segretin, M. E., Bos, J., Haas, B. J., Zody, M. C., Nusbaum, C., Win, J. O. E., Thines, M., & Kamoun, S. (2009). Ten things to know about oomycete effectors. Molecular Plant Pathology, 10, 795–803.
Silva, N. C., Nery, J. M., & Dias, A. L. T. (2014). Aspartic proteinases of Candida spp.: Role in pathogenicity and antifungal resistance. Mycoses, 57, 1–11.
Simões, I., & Faro, C. (2004). Structure and function of plant aspartic proteinases. European Journal of Biochemistry, 271, 2067–2075.
Sleebs, B. E., Lopaticki, S., Marapana, D. S., O'Neill, M. T., Rajasekaran, P., Gazdik, M., Günther, S., Whitehead, L. W., Lowes, K. N., Barfod, L., Hviid, L., Shaw, P. J., Hodder, A. N., Smith, B. J., Cowman, A. F., & Boddey, J. A. (2014). Inhibition of Plasmepsin V activity demonstrates its essential role in protein export, PfEMP1 display, and survival of malaria parasites. PLoS Biology, 12, e1001897.
Tarr, S. J., & Osborne, A. R. (2015). Experimental determination of the membrane topology of the Plasmodium protease Plasmepsin V. PLoS One, 10, e0121786.
Torto, T. A., Li, S., Styer, A., Huitema, E., Testa, A., Gow, N. A. R., van West, P., & Kamoun, S. (2003). EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Research, 13, 1675–1685.
Vleeshouwers, V. G. A. A., van Dooijeweert, W., Keizer, L. C. P., Sijpkes, L., Govers, F., & Colon, L. T. (1999). A laboratory assay for Phytophthora infestans resistance in various Solanum species reflects the field situation. European Journal of Plant Pathology, 105, 241–250.
Wang, S., Boevink, P. C., Welsh, L., Zhang, R., Whisson, S. C., & Birch, P. R. J. (2017). Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytology, 216, 205–215.
Wawra, S., Djamei, A., Albert, I., Nürnberger, T., Kahmann, R., & van West, P. (2013). In vitro translocation experiments with RxLR-reporter fusion proteins of Avr1b from Phytophthora sojae and AVR3a from Phytophthora infestans fail to demonstrate specific autonomous uptake in plant and animal cells. Molecular Plant Microbe Interactions, 26, 528–536.
Wawra, S., Trusch, F., Matena, A., Apostolakis, K., Linne, U., Zhukov, I., Stanek, J., Koźmiński, W., Davidson, I., Secombes, C. J., Bayer, P., & van West, P. (2017). The RxLR motif of the host targeting effector AVR3a of Phytophthora infestans is cleaved before secretion. The Plant Cell, 29, 1184–1195.
Whisson, S. C., Boevink, P. C., Moleleki, L., Avrova, A. O., Morales, J. G., Gilroy, E. M., Armstrong, M. R., Grouffaud, S., van West, P., Chapman, S., Hein, I., Toth, I. K., Pritchard, L., & Birch, P. R. J. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118.
Yaeno, T., & Shirasu, K. (2013). The RXLR motif of oomycete effectors is not a sufficient element for binding to phosphatidylinositol monophosphates. Plant Signaling & Behavior, 8, e23865.
Yamauchi, T., Nagahama, M., Hori, H., & Murakami, K. (1988). Functional characterization of Asp-317 mutant of human renin expressed in COS cells. FEBS Letters, 230, 205–208.
Zuluaga, A. P., Vega-Arreguín, J. C., Fei, Z., Ponnala, L., Lee, S. J., Matas, A. J., Patev, S., Fry, W. E., & Rose, J. K. C. (2016). Transcriptional dynamics of Phytophthora infestans during sequential stages of hemibiotrophic infection of tomato. Molecular Plant Pathology, 17, 29–41.
Acknowledgements
We acknowledge Dr. Justin Boddey for kindly providing the PMV-3HIS Plasmepsin V construct. This project was supported by the Food-for-Thought campaign from the University Fund Wageningen and by The Netherlands Organization for Scientific Research (NWO) in the framework of a VENI grant (Klaas Bouwmeester).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
Not applicable.
Informed consent
Not applicable.
Electronic supplementary material
ESM 1
(PDF 138 kb)
Figure S1
Pipeline for heterologous protein production in Escherichia coli. Details of protein production and purification are described in the materials and methods section. (PDF 69 kb)
Figure S2
P. infestans APs have enzymatic activity. In-gel zymography with soluble and refolded APs with 0.15% gelatin as substrate. Gels were stained with Coomassie Brilliant Blue to detect proteolytic activity as discoloration. Enzymatic assays were conducted in the absence (−) and presence (+) of the aspartic protease inhibitor pepstatin. (PDF 85 kb)
Figure S3
Alignment of full-length protein sequences of Phytophthora infestans APs and Plasmodium falciparum PMV. Signal peptide and propeptide regions are shown in italics and bold italics, respectively. Boxes indicate catalytic motifs with conserved aspartic acid (D) residues and the hydrophobic motifs (Kay et al. 2011). Amino acid positions highlighted in grey correspond to glutamic acid (E179) and glutamine (Q222) residues in PfPMV that are responsible for PEXEL substrate binding (Bedi et al. 2016). Cysteine (C) residues contributing to disulphide bonds are highlighted in black. C-terminal hydrophobic regions are underlined. The arrowhead points to the position corresponding to the alanine residue (A317) in human renin that contributes to a neutral pH optimum of this enzyme (Yamauchi et al. 1988). Symbols at the bottom line of the alignment indicate identical (*), strongly similar (:) and weakly similar (.) amino acid residues, respectively. (PDF 142 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Schoina, C., Verbeek-de Kruif, N., Govers, F. et al. Clade 5 aspartic proteases of Phytophthora infestans are virulence factors implied in RXLR effector cleavage. Eur J Plant Pathol 154, 17–29 (2019). https://doi.org/10.1007/s10658-019-01713-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01713-2