Abstract
Prune dwarf virus (PDV) is one of the most dangerous pathogens of fruit trees worldwide. One of the most important proteins required for PDV infection is replicase. (P1 protein) which anchored viral RNA and builds replication complex along with RNA depended polymerase. Despite the importance of PDV as a pathogen, our knowledge regarding tissue/cellular localization and structure of PDV P1 protein is still incomplete. The aim of this work was to localize replicase distribution in leaf tissues and cells by immunofluorescent and immunogold labeling of Nicotiana tobaccum cv Samsun and development of a 3D model of PDV replicase. In this paper we demonstrate that PDV replication, is similar to that of Alfalfa mosaic virus and is strongly connected with tonoplasts. In addition, PDV replicase and coat protein (CP) were also found to be strongly associated with membranes of endoplasmic reticulum and, indicating the potential involvement of these membrane structures in the processes related to viral infection. Bioinformatic analyzes based on 3D modeling and structure prediction revealed that P1 protein has a potential transmembrane domain which enables protein anchoring to tonoplast during replication complex assembly.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prune dwarf virus belongs to the family Bromoviridae and genus Ilarvirus (Fulton 1970; Fulton 1983; Szyndel and Paduch-Cichal 1997), i.e., the group of viruses with a segmented genome in the form of three RNA molecules (called RNA1, RNA2 and RNA3) packaged into separate particles. All three viral RNA components are required for infection of host cells (Bachman et al. 1994; Pallas et al. 2012; Kalinowska et al. 2014).
RNA1 encodes the replicase, also known as the P1 protein. This protein is involved in viral RNA anchoring in the site of assembly and functioning of the replication complex. RNA2 encodes the viral RNA-dependent RNA polymerase (RdRp) responsible for viral RNA replication (Codoñer et al. 2005, 2006). The PDV RNA3 encodes two proteins, i.e., movement protein (MP) and coat protein (CP). The MP is directly synthesized from RNA3 (Bachman et al. 1994; Pallas et al. 2012; Kozieł et al. 2015). The coat protein is synthesized from a-subgenomic RNA4 molecule (sgRNA4). In addition to its structural function, the PDV CP is involved in genome activation. Genome activation is a first step crucial for PDV replication. The N-terminal portion of Ilarvirus CP contains a large number of arginine (R) and/or lysine (K) residues, and is responsible for binding to the 3′UTR regions of viral RNA (Sánchez-Navarro and Pallas 1997). Ansel-McKinney et al. (1996) showed that arginine present in the N-terminal sequence of the protein is responsible for CP binding to RNA. The specialized domain binding RNA (RBD) with an affinity for the 3′UTR RNA3 and RNA4 operates in the CP of Prunus necrotic ring spot virus (PNRSV) which is closely related to PDV. This domain has a high arginine content and between amino acids 25 to 50 at the N-terminus of the protein, probably in all ilarviruses (Aparicio et al. 1999; Herranz et al. 2012; Kozieł et al. 2015). In members of the family Bromoviridae, the CP also has the ability to bind to the hairpin loop structure characteristic of the 3′UTR and initiate its transformation to the pseudoknot structure similar to that in transfer RNA-like structures (TLS) (Aparicio et al. 2003; Dreher 2009). The pseudoknot structure of viral RNA is necessary for recognition by RdRp in the replication complex. This structure is stabilized and maintained by the presence of Mg2+ (Aparicio et al. 2010). Magnesium ions inhibit the binding of further coat protein molecules, and also stimulate viral RNA binding to the replication complex (Dinant et al. 1993). Olsthoorn et al. (1999) and Bol (2005) suggested that P1 protein and coat protein are equally needed for replication of all Bromoviridae. The site of PDV genome replication in host plant cells is currently unknown. Therefore, the aim of this study was to determine the tissue and subcellular distribution of two proteins involved in the replication process, i.e., coat protein, responsible for the process of genome activation, and replicase which is co-involved in formation of the replication complex and anchoring of viral RNA to plant cell membranes. Using immunolabeling methods we demonstrated that PDV CP and P1 protein strongly localized in tonoplast but also in membranes of endoplasmic reticulum and chloroplasts of infected tobacco cells. Our results indicated that PDV CP and P1 are connected with different membrane structures in infected cell and its localization/deposition strongly determined replication process areas in plant cell. Moreover, the 3D model of replicase structure and analyzes by use of TMHMM and ΔG Prediction server showed that P1 protein has a potential transmembrane domain that enables protein anchoring to the tonoplast during replication complex assembly.
Materials and methods
PDV inoculation
The research material for analysis of replicase and coat protein PDV distribution were tobacco plants (Nicotiana tabacum cv. Samsun). Tobacco plants were inoculated mechanically with Carborundum using leaf tissue extracts of plum infected with PDV isolate 0599. Tissue extraction buffer was 0.01 M potassium-phosphate buffer (pH 7.2–7.4) containing Na-DIECA (according to Waterworth and Fulton 1964). The virus source was obtained from the Institute of Horticulture in Skierniewice, due to courtesy of Tadeusz Malinowski, PhD. The plants were grown under phytotronic conditions, at 18 °C and 16-h light with an intensity of 400 μmol m−2 s−1 PAR (photosynthetically active radiation).
Material preparations for immunofluorescent localization of PDV coat protein (CP) and replicase (P1 protein)
Samples of tobacco leaf blades were collected 15 days after inoculation from plants infected with PDV-0599 and from healthy plants, washed in 0.1 M MSB buffer (pH 6.9) then fixed in 4% (w/v) paraformaldehyde on 0.1 M MSB (pH 6.9), dehydrated in ethanol with DTT and supersaturated with BMM (butyl-methyl- acrylate) resin (Sigma-Aldrich, Germany) (according with procedure presented by Barthels et al. 1997; Otulak et al. 2016). The material embedded in resin was polymerized for 20 h at −20 °C using UV lamps (Rafińska et al. 2014).
Immunofluorescent localization of coat protein (CP) and replicase (P1 protein)
Fragments of tobacco leaves embedded in BMM were analyzed both after infection with PDV and in buffer mock-inoculated plants used as controls. Immunolocalization was performed separately for P1 and CP. A series of macro-sections of leaf blades fragments of a thickness of 2–3 μm were made using glass knives on RM 2065 microtome (Reichert-Jung). The sections were placed on a silylated slide (Thermo) in drops of water as described by Otulak et al. (2016) with one modification. Sections were glued on thermoblocks at 45 °C by 24 h. Next steps in procedure (acetone and distilled water washing, slide preparation, PBS pre-incubation and PBS-BSA blocking) were performed according to method already presented in Otulak et al. (2016). The slides were then washed in PBS, incubated for one hour with primary purified rabbit polyclonal antibody anti-CP-PDV (Bioreba, Switzerland) or P1-PDV (detecting TRQVGPKAQYERGYTVN fragment of P1 protein of the reference isolate PDV-Salmo BC cherry, NCBI Protein: YP_611154.1, GeneCust, Luxemburg) in dilution of 1: 100 and 1:50, respectively, in PBS. Four rinses in 0.01 M PBS buffer with 0.05% Tween 20, and then one rinse in 0.01 M PBS were applied. The slides were then treated with secondary anti-rabbit antibody IgG 1:100 coupled to a fluorescent dye FITC (fluorescein, Agrisera, Sweden) for 1 h in the dark. After incubation the sections were washed by PBS-Tween and PBS and stained by DAPI (4′, 6-diamidino-2-phenylindole). The observations were carried out by use of light-fluorescent microscope (Otulak and Garbaczewska 2010; Otulak et al. 2016). Parallel to the immunolabeling, the tests using double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) were performed to verify the specificity of P1 antibodies for virus detection. The above-mentioned anti-CP-PDV antibodies (a standard for PDV detection in plant material (Clark and Adams 1977; Abou-Jawdah et al. 2004) and, separately, an anti-P1-PDV antibody were used. Both tests gave similar results of absorbance measurement at a wavelength of 405 nm at a level of 0.60 for CP and 0.4 for protein P1. As suggested by Paduch-Cichal (2000) and Paduch-Cichal and Sala-Rejczak (2003) absorbance values above 0.2 was the confirmation of virus detection.
Immunolocalization of coat protein (CP) and replicase (protein P1) using the antibodies with colloidal gold
Analysis involved the fragments of tobacco leafs after infection caused by PDV and control (mock-inoculated) plants. Material was fixed, dehydrated and embedded in accordance with the procedure presented by Otulak et al. (2016). Ultrathin sections (90–110 nm) were cut on an ultramicrotome (Reichert-Jung Ultracut E), transferred to nickel grids with formvar, and then subjected to labeling (Hayat 1986). The grids were treated according to procedure presented by Otulak and Garbaczewska (2010). Then, they were washed in 0.01 M PBS-Tween 20 incubated for one hour in primary antibodies as for immunofluorescence. Rabbit anti-CP-PDV antiserum was used at 1:100 and anti-P1-PDV antiserum at 1:50 in PBS buffer. After rinsing in 0.01 M PBS-Tween 20, the grids were incubated with secondary anti-rabbit IgG conjugated with colloidal gold (20 nm) for 1 h (1:100 for CP-PDV labeling, and 1:50 for P1-PDV labeling). The grids were rinsed, stained and then observed by transmission electron microscopy (Otulak and Garbaczewska 2010).
Predicting the spatial structure of PDV replicase (P1 protein)
An identification of the key active domains and fragment to which the antibodies were designed, involved the use of AIDA server (Ab inito domain assembly server) (http://ffas.burnham.org/AIDA/), in which the model of surface and tertiary structure of P1 protein was created based on the reference sequence of isolate PDV-Salmo BC cherry (NCBI protein: YP_611154.1).
In the initial stage after the complete sequence of protein introduction in the FASTA format, PSI-BLAST program searches the sequence for the single domains based on a databases containing known protein structures. After determination of the existing domains, PSIPRED algorithm provides the secondary structures present in the domains(Xu et al. 2014). In the next step, AIDA predicts the lengths, angles between the bonds and torsion angles between residues in the sequence. In the final stage, the ab into algorithm assembles single domains based on PSI-Blast, PSIPRED data and the information about the bonds, and forecasts the probable protein folding (Xu et al. 2015). The structures obtained in AIDA were then further developed using programs PDBViewer (http://spdbv.vital-it.ch/) and PyMol (https://www.pymol.org/) to show the protein surface and the spatial arrangement of secondary structures (Guex and Peitsch 1996; Guex et al. 2009).
Analysis of the spatial structure of PDV replicase (P1 protein) for the presence of transmembrane structures
Two bioinformatics programs: TMHMM (Sonnhammer et al. 1998) and ΔG Prediction Server 1.0 (Martínez-Gil et al. 2009) were used to verify whether P1 protein belonging to PDV forms a transmembrane domain. The TMHMM program searches the protein sequence for strongly hydrophobic fragments, which may form transmembrane domains. From the hydrophobicity of certain regions of the sequence, TMHMM calculates the probability and location of the transmembrane domain (Krogh et al. 2001; Moller et al. 2001). An ΔG Prediction 1.0 server calculates apparent free energy difference of bonds between amino acids (ΔGapp), and on that basis determines the site of transmembrane domain occurrence. In most cases, the negative ΔGapp value indicates the presence of transmembrane domain.
Results and discussion
Immunofluorescent and immunogold labeling localization of CP-PDV in leaf tissues
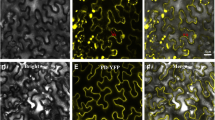
Fifteen days after PDV inoculation, labeling was observed in most tissues of the leaf blade. Green fluorescent signal derived from CP-PDV epitopes was observed in palisade and spongy parenchyma cells (Fig. 1a, b), but also in necrotic phloem cells (Fig. 1c) and tracheal xylem elements (Fig. 1d). Moreover, cell wall invaginations were visible in palisade and spongy parenchyma cells with CP location (Fig. 1a, b). No labeling or anatomical lesions related to infection were noted in tobacco plants inoculated with buffer (mock-inoculated) (Fig. 1e).
Immunofluorescent localization of coat protein in tobacco leaf of Samsun variety 15 days after inoculation with PDV. a Cross-section of tobacco leaf blade. The epitopes of coat protein (*) visible in palisade and spongy parenchyma cells. Magnification of leaf fragment in Fig. B (white frame). b Magnification of figure a. The epitopes of coat protein (*) visible in phloem cells, palisade and spongy parenchyma. Disintegration of spongy parenchyma cells observed (arrow). c The epitopes of coat protein (*) in necrotic phloem cells. Visible cell wall invagination of parenchyma cells (arrow). d The epitopes of coat protein (*) in the spongy parenchyma cell and tracheal element. e Fragments of leaf blades inoculated only with buffer. Cross section of tobacco leaf 15 days after inoculation with buffer. No CP epitopes location. Abbreviations: Ep-epidermis, PMe-palisade parenchyma, SMe-spongy parenchyma, VB-vascular bundle, X- tracheal element of xylem, Ph-phloem, Ne-necrosis
Immunofluorescent localization of CP-PDV showed that after inoculation of tobacco plants with PDV, viral coat protein is present in the same cells in which the virus particles were observed. The fluorescent signal derived from CP-PDV epitopes was observed in the mesophyll cells of both types, but also within the phloem. A similar relationship was observed in the cells of Nicotiana benthamiana infected with Alfalfa mosaic virus (AMV) (van Pelt-Heerschap et al. 1987; van der Wel et al. 1998). Seven days after inoculation with AMV, only the cells of palisade and spongy parenchyma were subject to the labeling (van der Wel et al. 1998). Further observations regardless of the kind/model of immunolabeling showed that CP-PDV localized in most tobacco tissues of the leaf blade. Such high level of CP distribution are connected with infection spread and involvement of CP. Pallas et al. (2012) showed that coat protein quantitatively dominates over other viral proteins in plant cells infected by the representatives of Bromoviridae family. Bol (2005) suggests that the amount of coat protein is a result of its involvement during the viral infection at the early stages. CP is involved in genome activation, asymmetric accumulation of (+) strand RNA, translation of viral RNA. Formation of virions and interstitial and systemic transport is also depended on CP. RNA strands in the Bromoviridae family, which includes PDV, are synthesized asymmetrically, approximately 100 molecules (+) accounts for one (−) (Houwing et al. 1998).
Transmission electron microscopy observation revealed the location of the antigen of C-terminal CP-PDV fragment. The presence of colloidal gold particles was observed in many mesophyll cell organelles. CP-PDV epitopes were strongly localized in the vacuole (especially in tonoplast), in the mitochondria and chloroplasts, in the cytoplasm, and also near the plasmodesmata (Fig. 2a, b). As for immunofluorescence location, the presence of antigen of the C-terminal CP-PDV fragment was confirmed in phloem. CP-PDV was located inside the sieve tubes and companion cells (Fig. 2c, d). In turn, in the companion cells, CP PDV epitopes were observed near cell organelles. CP-PDV was noted in the membranes of vesicles present in the cytoplasm (Fig. 3a, b) and inside the vacuole (Fig. 3c), and even in the nucleus (Fig. 3a). Moreover, in parenchyma cells CP-PDV epitope was also observed in spherules membranes (Fig. 3d, e). Inside spherules PDV particles presence was confirmed (Fig. 3e). Inside companion cells, viral particles and expanded cisternae of endoplasmic reticulum were labeled (Fig. 3a, c). Decoration with colloidal gold particles was not limited to the phloem, the location of CP antigen was often observed in tracheal elements of xylem and in vacuoles of wood parenchyma cells (Fig. 3f).
Immunolocalization of CP-PDV using antibodies with colloidal gold in the tissues of leaf blade of Samsun variety tobacco 15 days after inoculation with PDV. a Numerous colloidal gold particles (*) in chloroplast and protoplast of mesophyll cell. b Colloidal gold particles (*) in vacuole of mesophyll cell. Labeling was observed in cytoplasm (arrow) and by plasmodesmata. c Colloidal gold particles (*) in mesh tube protoplast. Vesicles separating from plasma membrane visible in sieve tube. d Numerous colloidal gold particles (*) in sieve tube. Labeling was observed in plasma membranes between sieve tube and companion cell. Abbreviations: CW-cell wall, Ch-chloroplast, St-starch, VP-viral particles, V-vacuole, vs-vesicle, PD-plasmodesmata. M-mitochondrion, PLs-plastid, CC-companion cell, SE-sieve tube
Immunolocalization of PDV coat protein (CP) using antibodies with colloidal gold in the tissues of leaf blade of Samsun variety tobacco 15 days after inoculation with PDV. a Labeling was observed in nucleus (N) and on ER surface (*). b Colloidal gold particles (*) in companion cell and sieve tube. Labeling was observed in nucleus (N) of phloem parenchyma cell. c Colloidal gold particles (*) in companion cells protoplast. Labeling was observed in vesicles and vacuole of companion cells. d Colloidal gold particles (*) in spherules membrane in parenchyma cell. White frame area in Fig. 3e. e Enlarge fragment with spherules in white frame from Fig. 3d. f Colloidal gold particles (*) in xylem tracheal element and wood parenchyma. Abbreviations: CW-cell wall, Ch-chloroplast, M-mitochondrion, ER-endoplasmic reticulum, VP-viral particles, vs-vesicle, CC- companion cell, PP-phloem parenchyma, SE- sieve tube, Sp-spherule, X- tracheal element of xylem, XP- xylem parenchyma
Colloidal gold particles indicating the presence of CP PDV were localized in a number of cellular organelles and near the PDV particles present in infected cells. The labeling pattern was associated with vacuole, tonoplast, chloroplasts and endoplasmic reticulum. The presence of CP-PDV in vacuoles is closely related to PDV virus particles observed there. It appears that the vacuole is a site of PDV particle assemblage, as observed for AMV (Ibrahim et al. 2012). Moreover, van der Wel et al. (1998) observed the labeling of CP AMV in vacuoles of tobacco cells filled with or without AMV particles. Van der Hejden et al. (2001) and Ibrahim et al. (2012) noted the presence of CP near tonoplast of infected protoplasts of Arabidopsis thaliana. The same authors showed that the tonoplast is the site of viral RNA (vRNA) AMV replication. Presence of CP in tonoplast after genome activation is an effect of asymmetric vRNA accumulation (Bol 2005). Olsthoorn et al. (1999) proposed a model for a conformational switch affecting the accumulation of sense and antisense strands on the example of AMV and may be also characteristic for PDV. In this model, CP attaches to the 3′ end preventing the formation of pseudoknot structure, and thus makes that RdRp is unable to recognize the RNA used in the synthesis of (−) strand, promoting the asymmetric synthesis of (+) strand. The promoter of the initiation of (−) strands transcription are AUG loops in 3′UTR TLS of (+) strands, while for the sense strand it is the 3′ end of the antisense strand (Sivakumaran et al. 2000; Sivakumaran et al. 2002). Localization of CP-PDV in ER indicates the site of this protein synthesis and may suggest its potential involvement in the transmission of viral RNA used in the translation. This seems to be probable, because PDV coat protein, like that of other Ilarviruses, has the domain capable of RNA binding (Pallas and Garcia 2011; Pallas et al. 2012; Pallas et al. 2013). In the case of AMV, there are no reports concerning CP location in the ER, which seems to indicate the difference between this virus and PDV. Nevertheless, a strong labeling of vesicles and ER was showed in case of Brome mosaic virus (BMV), another member of the Bromoviridae family (Bamunusinghe et al. 2011). Sztuba-Solińska and Bujarski (2008) showed that the sites of BMV replication are ER membranes, and the vesicles differentiating from the ER membrane can be a potential site of particles accumulation. BMV creates special membrane structures-spherules in which replication occurs. A strong labeling of mitochondria was observed in PDV infected tobacco plants. No location of CP in mitochondria was noted both for AMV and BMV (van der Hejden et al. 2001; Bamunusinghe et al. 2011). Moreover, the locations of coat protein epitopes were observed in the area of chloroplasts. Similar locations in chloroplast, but also in other parts of the cell such as the nucleus or nucleolus, were observed for AMV (van Pelt-Heerschap et al. 1987). The changes in chloroplasts were observed during AMV-induced infection, in which the viral particles were visible over the time (Hull et al. 1969a; Hull et al. 1969b; Favali and Conti 1970). Favali and Conti (1970) and van Pelt-Heerschap et al. (1987) demonstrated the presence of CP and AMV viral particles inside chloroplast, therefore based on these microscopic findings they postulated that replication process may be connected with this organelles.
Immunofluorescent and immunogold labeling localization of PDV replicase in leaf tissues
The presence of an antigen of PDV P1 protein was shown in the tissues of tobacco leaves 15 days after inoculation in protoplasts of palisade and spongy parenchyma cells (Fig. 4a), as well as in necrotic phloem cells (Fig. 4b).
Immunofluorescent localization of P1 protein (replicase) in tobacco leaf of Samsun variety 15 days after inoculation with PDV. a Cross-section of tobacco leaf blade. The epitopes of P1 protein (*) visible in palisade and spongy parenchyma cells. b Epitopes of P1 protein (*) in parenchyma. Necrotic changes in phloem. c Cross section of tobacco leaf 15 days after inoculation with buffer. No locations of P1 protein epitopes. Abbreviations: Ep-epidermis, PMe-palisade parenchyma, SMe-spongy parenchyma, Pa-parenchyma, X-tracheal element, Ph-phloem, Ne-necrosis
Immunogold labeling revealed by TEM the presence of antigen of C-terminal fragment of P1 protein, like CP, in mesophyll cells (Fig. 5a, b). In spongy parenchyma cells, epitopes of protein P1 were localized within the vacuole, chloroplasts, as well as vesicles (Fig. 5b, c). The location of replicase in chloroplasts and mitochondria was often accompanied by changes in their ultrastructure. Chloroplasts were subject to deformation (Fig. 5b), and electron-translucent regions were observed in mitochondria (Fig. 5d). Moreover, also in parenchyma cells spherules near tonoplast were labeled (Fig. 5e, f). Within the phloem, strong decoration with colloidal gold particles was observed in the vacuoles of companion cells, phloem parenchyma cells and inside the sieve tubes (Fig. 6a, c). The presence of an antigen of C-terminal fragment P1-PDV was observed in the vacuoles of companion cells, which cytoplasm was filled with PDV virus particles (Fig. 6b, c). Often, the epitopes of P1 protein in phloem parenchyma cells were localized in the endoplasmic reticulum (Fig. 6d).
Immunolocalization of P1 protein (replicase) using antibodies with colloidal gold in mesophyll cells of tobacco leaf of Samsun variety 15 days after inoculation with PDV. a Gold particles (*) in spongy parenchyma cells vacuoles. Magnification of the fragment of spongy parenchyma cell cytoplasm in Fig. 5b (white frame). b Magnification of Fig. 5a. Epitopes of P1 protein (*) visible in chloroplast and vesicles of spongy parenchyma cells. c Gold particles (*) in vacuoles, vesicles and chloroplasts of palisade parenchyma cell. d Colloidal gold particles in palisade parenchyma cell (*) visible in chloroplast, vacuole and in vicinity of mitochondria with electron-translucent area. e Gold particles (*) in parenchyma cell tonoplast and in membranes of spherules. White frame area in Fig. 5f. f Enlarge fragment with spherules in white frame from Fig. 3e. Abbreviations: CW-cell wall, Ch-chloroplast, ER- endoplasmatic reticulum, V-vacuole, vs-vesicle, M-mitochondrion, SMe-spongy parenchyma, Sp-spherule, PD-plasmodesmata
Immunolocalization of P1 protein (replicase) using antibodies with colloidal gold in companion cells of Samsun variety tobacco 15 days after inoculation with PDV. a Epitopes of P1 protein (*) visible in vacuoles of companion cell. Viral particles visible in companion cell. Magnification of companion cell in Fig. 6b (white frame). b Colloidal gold particles (*) visible in companion cell vacuoles. c Epitopes of P1 protein (*) visible in vacuoles of phloem parenchyma and companion cells, and inside sieve tubes. Viral particles visible in companion cell. d Colloidal gold particles (*) visible on endoplasmic reticulum surface and inside phloem parenchyma cell vacuole. e Fragments of vascular bundle of tobacco inoculated with PDV without primary antibody. Abbreviations: CW-cell wall, PD-plasmodesmata, N-nucleus, M-mitochondrion, ER-endoplasmic reticulum, V-vacuole, VP-viral particles, vs-vesicle, SE- sieve tube. PP-phloem parenchyma, CC-companion cell
Labeling was not observed either in control tobacco plants inoculated with buffer alone (Fig. 4c), or in samples of tobacco plants inoculated with PDV without primary antibody (Fig. 6e).
PDV replicase (P1 protein) is probably involved in viral RNA anchoring during replication (Pallas et al. 2013). In case of all Bromoviridae, this protein co-localizes with RNA-dependent polymerase RNA (P2 protein) in the postulated areas of replication (Bol 2005; Pallas et al. 2012). The process of PDV RNA synthesis requires the formation of a replication complex composed of P1 and P2 proteins, within the appropriate cell organelles (Bol 2005; Pallas et al. 2013). The results of fluorescent and immunogold labeling suggested a similar location of P1 protein and CP of PDV virus in certain tissues. Replicase was localized identically as CP PDV around both types of tobacco leaf blade mesophyll, phloem parenchyma, sieve tubes necrotic altered phloem cells. The observations using TEM also showed some similarities and minor differences between the subcellular distribution of P1 protein and CP. Particles of colloidal gold indicating the epitopes of P1 protein were observed in mitochondria, chloroplasts, ER, moreover, also in vacuoles, and tonoplast within parenchyma cells. Bamunusinghe et al. (2011) pointed out that within Bromoviridae family, the coat protein is near the sites of occurrence of P1 and P2 protein (replication complex). Such close presence of CP and replication complex is closely related to the need to change the conformation of viral RNA, i.e., so-called genome activation and also with asymmetric RNA replication determined by CP (van der Vossen et al. 1994; Bol 2005; Pallas et al. 2013). PDV P1 protein was strongly localized in the membranes of various types of cell organelles and in spherules membranes which is partly consistent with the literature (Bamunusinghe et al. 2013; Bol 2005; Pallas et al. 2012; Pallas et al. 2013). In Bromoviridae family, P1 protein is associated with various membrane structures and its location is determined by the site of replication. BMV P1 protein is localized in ER, where the virus induces the process of replication and formation of various kinds of vesicles and spherules (Liu et al. 2009; Bamunusinghe et al. 2013). Spherules are membranous structures appear during many Bromoviridae viral infections indicates ongoing RNA replication (Bol 2005). Bamunusinghe et al. (2013) showed in infections of BMV and Cucumber mosaic virus (CMV) that spherules may be membranous invagination or spheral membranous structures respectively near ER (in case of BMV) or tonoplast (in case of CMV). Therefore, presence of viral particles, PDV-CP and P1 in spherules during infection definite indicates replication process. AMV and CMV replication occurs in the cells tonoplast and the particles are often accumulated in the vacuole (van der Hejden et al. 2001; Cillo et al. 2002). Ibrahim et al. (2012) confirmed not only the location of the AMV P1 and P2 proteins, but also viral RNA in tonoplast and vacuole. However, there have been no data so far on the location of P1 protein in organelles such as vacuole, mitochondria and chloroplasts. The presence of the virus and viral proteins in organelles can have two possible explanations. The first indicates that PDV and AMV could in a certain range use the translation apparatus of semi-autonomous organelles, and the replication of the viral RNA may involve outer membranes of chloroplasts and mitochondria. The second one, more likely, indicates that CP, P1 protein could be transported to this organelles by another unknown mechanism. The second hypothesis is supported by observations from immunocytochemical labeling. The presence of P1 protein in the vesicles observed in certain locations within the plant, suggest that this protein can be transported in the cell in those membranous structures.
In conclusion, the immunocytochemical labeling of P1 PDV protein demonstrates that its replication, as AMV, is primarily related to the tonoplast.
Predicting secondary and tertiary structures and analysis of the spatial structure of PDV replicase (P1 protein) for the presence of transmembrane domains
Analysis of the amino acid sequence of P1 protein of reference isolate PDV-Salmo BC cherry by using the programs on the AIDA server permitted the creation of a model of the tertiary structure. Two characteristic domains located at opposite ends of the P1 protein were detected. A methyltransferase RNA domain (orange frame in Fig. 7a) located between MET1 and SER471 comprising 471 amino acids was identified at the N-terminus. The methyltransferase domain consisted of α-helical regions separated by regions of simple polypeptide chain (Fig. 7b), and regions forming the structure of β-card (marked in yellow in Fig. 7b). The predicted helicase domain was detected at the C-terminus of protein P1 (white frame in Fig. 7a), which included 457 amino acids, and was localized between ALA598 and VAL1055. It structural composition was similar to methyltransferase and consisted of α-helical, β-card and simple polypeptide chain regions (Fig. 7c). The helicase domain between THR692 and ASN708 contained marked in red highly antigenic fragment, which was detected during the immunolabeling (Fig. 7c). For better understanding of protein function visualization of protein surface (Fig. 8a) and individual domains were created in PDB Viewer (Fig. 8b, c).
3D structure of PDV replicase (P1 protein) made in AIDA program. Three-dimensional structure of the entire P1 protein with marked domains. a The colors showed particular elements of secondary structure. Green indicates the fragments of straight polypeptide chain, blue α-helical fragments, yellow β-card fragments. Orange frame- methyltransferase domain, white frame- helicase domain. Buckle shows helical region between domains. b Magnification of methyltransferase domain from Fig. 7a. Visible clockwise (*) and anticlockwise (arrow) α-helical regions. Green indicates a simple polypeptide chain, yellow fragments of β-card structure. c Magnification of helicase domain (from Fig. 7a) rotated 90 degrees to the vertical axis with marked in red fragment used in P1 protein immunolocalization. Also visible clockwise (*) and anticlockwise (arrow) α-helical regions. Green indicates a simple polypeptide chain, yellow fragments of β-card
3D structure of PDV replicase (P1 protein) considering surface and transmembrane domain. The spatial structure of the entire P1 protein with marked domains. a The colors showed the particular elements of secondary structure. Green indicates the fragments of straight polypeptide chain, blue α-helical fragments, yellow β-card fragments. Orange frame- methyltransferase domain, white frame-helicase domain. Buckle shows helical region between domains. Protein surface applied to its 3D structure structure is marked in gray. b. Magnification of. methyltransferase domain from Fig. 8a. Gray indicates protein surface. c Magnification of helicase domain (from Fig.8a) rotated 90 degrees to the vertical axis with marked in red fragment used in P1 protein immunolocalization. Purple indicates probable transmembrane domain. The area of helicase domain applied to its 3D structure is marked gray
The analysis using THMM program allowed the detection of one probable transmembrane domain, which occurred between amino acids 848–869. The transmembrane sequence AGVIYAAATIAGVSEVIAFGDT was in the helicase domain. Further analysis using the PDB Viewer program confirmed that the secondary structure of this fragment is α-helical (Fig. 8c). The probability of this domain occurrence estimated by THMM was about 0.1, i.e., 10% (Fig. 9a). Full analysis of protein sequence made using ΔG Prediction Server 1.0 showed the same sequence fragment located between amino acids 848–869 (Fig. 9b). Apparent free energy difference calculated for the sequence fragment (ΔGapp) was +2.574 kcal/mol (Fig. 9b).
Analysis of transmembrane domains of PDV P1 protein. a Diagram showing the location of transmembrane domain and the likelihood of its occurrence. Red indicates transmembrane fragment (arrow), blue intracellular domain, pink extracellular domain. Diagram made in THMM program. b Diagram showing free energy calculated by ΔG Prediction Server 1.0 for P1protein. Gray stripe indicates observed transmembrane domain
Predicting of the tertiary structure of PDV P1 protein was conducted for the first time, since applied AIDA Server has been operating only since 2014. Therefore, there was no literature describing this type of modeling for P1 protein. N-terminal methyltransferase domain is responsible for zero type cap attaching to the 5′ end of mRNA, preventing thus RNA exonucleases activity present in the host organism which destroy unprotected RNA (Rozanov et al. 1992). Complex structure of this domain (numerous α-helical and β-card regions) is consistent with the results of bioinformatic analysis of P1 AMV, BMV and CMV protein sequences obtained by Rozanov et al. (1992), and is also compatible with the characteristics of these types of domains of Ilarvirus genus representatives (Pallas et al. 2012). C-terminus UvrD/REP helicase domains, breaks the hydrogen bonds between the nucleic acids during the replication (Rozanov et al. 1992; Korolev et al. 1997; Ramptish and Estewell 1997). Koonin and Dolja (1993) and Korolev et al. (1997) leading the sequence analysis suggested the presence of α-helical and β-card regions within helicase domains of AMV and CMV. Both domains were separated by the fragment of a clockwise α-helix including 123 amino acids (between TYR 472 and ALA 594). P1 protein is involved in the capture of viral RNA in the sites of replication complex accumulation (Bol 2005; Pallas et al. 2012). The presence of such long fragment between the domains indicates a site of the location of the second element of the replication complex encoded by RNA2, i.e., RNA-dependent polymerase RNA (P2 protein). The studies concerning BMV and AMV replication show the co-localization of both P1 and P2 proteins within the ER or tonoplast (Schwartz et al. 2002; Schwartz et al. 2004; Ibrahim et al. 2012). This type of both proteins co-localizating may indicate that P1 and P2 proteins may be connected during formation and functioning of the replication complex, and the site of their association may be an inter-domain region of P1 protein.
The results regarding immunocytochemical localization of PDV P1 protein showed its location in membrane structures. Moreover, literature data indicate the location of P1 protein of BMV and AMV viruses in ER membranes or tonoplast (Schwartz et al. 2004; Ibrahim et al. 2012). Thus, there was the question whether this protein is capable of anchoring in a membrane being a transmembrane protein, or perhaps there is another factor that allows location in membrane structures of the cell during infection. A deeper analysis of the sequence of P1 protein from PDV-Salmo BC isolate with THMM program allowed to detect one probable transmembrane helicase domain. Further analysis in the PDB Viewer confirmed that the secondary structure of this fragment is α-helical, which would enable the formation of transmembrane domain. The size of probability estimated by THMM did not allow to confirm unequivocally the presence of this type of domain. Therefore, the analysis was extended by the difference in free energy based on ΔG Prediction Server 1.0. The results showed the same part of the P1 sequence. Apparent free energy difference calculated for the fragment of the sequence difference (ΔGapp) was +2.574 kcal/mol. In order to confirm the existence of the domain, the free energy difference should be negative (Hessa et al. 2005). However, when the value of polypeptide bonds energy have a small positive value (1–3 kcal/mol), the possible existence of transmembrane domains is determined by the presence of α-helical structures in the spatial vicinity of the potential transmembrane domain (Hessa et al. 2005; Hessa et al. 2007). The structures of this type stabilize the transmembrane structure and enable its correct location in the membrane (Hessa et al. 2007; Sonnhammer et al. 1998). Four α-helical domains were identified in P1 protein in the surrounding of selected amino acid residues, which could potentially stabilize the protein structure. So far, there is no data in the literature on the presence of transmembrane domains in P1 protein. In turn, Martínez-Gil et al. (2009) confirmed using THMM and ΔG Prediction Server 1.0, the presence of domain allowing the integration with cell membranes at the C-terminus of PNRSV movement protein. The presence of this domain probably enables the transport of viral RNA bound by RBD to specific membrane structures (Martínez-Gil et al. 2009). The analysis of transmembrane structures confirmed the membrane location of PDV P1 protein. It seems that due to the presence of transmembrane domain, this protein can locate in tonoplast or membranes of other organelles in the cells of infected host plants.
References
Abou-Jawdah, Y., Sobh, H., Cordahi, N., Kawtharani, H., Nemer, G., Maxwell, D. P., & Nakhla, M. K. (2004). Immunodiagnosis of Prune dwarf virus using antiserum produced to its recombinant coat protein. Journal of Virological Methods, 121, 31–38.
Ansel-McKinney, P., Scott, S. W., Swanson, M., Ge, X., & Gehrke, L. (1996). A plant viral coat protein RNA binding consensus sequence contains a crucial arginine. EMBO Journal, 15, 5077–5084.
Aparicio, F., Sánchez-Navarro, J. A., & Pallás, V. (2010). Implication of the C terminus of the Prunus necrotic ringspot virus movement protein in cell-to-cell transport and in its interaction with the coat protein. Journal of General Virology, 91, 1865–1870.
Aparicio, F., Sánchez-Pina, M. A., Sánchez-Navarro, J. A., & Pallás, V. (1999). Location of Prunus necrotic ringspot ilarvirus within pollen grains of infected nectarine trees: Evidence from RT-PCR, dot-blot and in situ hybridisation. European Journal of Plant Pathology, 105, 623–627.
Aparicio, F., Vilar, M., Perez-Paya, E., & Pallas, V. (2003). The coat protein of Prunus necrotic ringspot virus specifically binds to and regulates the conformation of its genomic RNA. Virology, 313(1), 213–223.
Bachman, E. J., Scott, S. W., Xin, G., & Vance, V. B. (1994). The complete nucleotide sequence of prune dwarf ilarvirus RNA 3: implications for coat protein activation of genome replication in ilarviruses. Virology, 201(1), 127–131.
Bamunusinghe, D., Chaturvedi, S., Seo, J. K., & Rao, A. L. (2013). Mutations in the capsid protein of Brome mosaic virus affecting encapsidation eliminate vesicle induction in planta: Implications for virus cell-to-cell spread. Journal of Virology, 87(16), 8982–8992.
Bamunusinghe, D., Seo, J. K., & Rao, A. L. (2011). Subcellular localization and rearrangement of endoplasmic reticulum by Brome mosaic virus capsid protein. Journal of Virology, 85(6), 2953–2963.
Barthels, N., van der Lee, F. M., Klap, J., Goddjin, O. J. M., Karimi, M., et al. (1997). Regulatory sequences of arabidopsis drive reporter gene expression in nematode feeding structures. The Plant Cell, 9, 2119–2134.
Bol, J. F. (2005). Replication of alfamo- and ilarviruses: Role of the coat protein. Annual Review of Phytopathology, 43, 39–62.
Cillo, F., Roberts, I. M., & Palukaitis, P. (2002). In situ localization and tissue distribution of the replication-associated proteins of cucumber mosaic virus in tobacco and cucumber. Journal of Virology, 76, 10654–10664.
Clark, M. F., & Adams, A. N. (1977). Characteristics of the microplate method of ELISA for the detection of plant viruses. Journal of General Virology, 34, 475–483.
Codoñer, F. M., Cuevas, J. M., Sánchez-Navarro, J. A., Pallas, V., & Elena, S. F. (2005). Molecular evolution of the plant virus family Bromoviridae based on RNA3-encoded proteins. Journal of Molecular Evolution, 61, 697–705.
Codoñer, F. M., Fares, M. A., & Elena, S. F. (2006). Adaptive covariation between the coat and movement proteins of Prunus necrotic ringspot virus. Journal of Virology, 80(12), 5833–5840.
Dinant, S., Janda, M., Kroner, P. A., & Alquist, P. (1993). Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. Journal of Virology, 67, 7181–7189.
Dreher, T. W. (2009). Role of tRNA-like structures in controlling plant virus replication. Virus Research, 139(2), 217–229.
Favali, M. A., & Conti, G. G. (1970). Ultrastructural observations on the chloroplasts of basil plants either infected with different viruses or treated 3-amino-1,2,4-triazol. Protoplasma, 70, 153–166.
Fulton, R.W. (1970). Prune dwarf virus. C.M.I/a.A.B. Description of Plant Viruses, 19.
Fulton, R.W. (1983). Ilavirus group. C.M.I/a.A.B. Description of Plant Viruses, 274.
Guex, N., & Peitsch, M. C. (1996). Swiss-PdbViewer: A fast and easy-to-use PDB Viewer for Macintosh and PC. Protein Data Bank Quaterly Newsletter, 77, 7–10.
Guex, N., Peitsch, M. C., & Schwede, T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis, 1, 163–171.
Hayat, M. (1986). Basic techniques for transmission electron microscopy. New Jersey, Academic Press International.
Herranz, M. C., Pallas, V., & Aparicio, F. (2012). Multifunctional roles for the N-terminal basic motif of Alfalfa mosaic virus coat protein: Nucleolar/cytoplasmic shuttling, modulation of RNA-binding activity, and virion formation. Molecular Plant-Microbe Interactions, 25, 1093–1103.
Hessa, T., Kim, H., Lundin, C., Boekel, J., Andersson, H., Nilsson, I., White, S. H., & von Heijne, G. (2005). Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature, 433, 377–381.
Hessa, T., Meindl-Beinker, N., Bernsel, A., Kim, J., Sato, Y., Lerch, M., Lundin, C., Nilsson, I., White, S. H., & von Heijne, G. (2007). Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature, 450, 1026–1030.
Houwing, C. J., van de Putte, P., & Jaspars, E. M. J. (1998). Regulation of single strand RNA synthesis of Alfalfa mosaic virus in non-transgenic cowpea protoplasts by the viral coat protein. Archives of Virology, 143, 489–500.
Hull, R., Hill, G. J., & Markham, R. (1969a). Studies on Alfalfa mosaic virus: II. The structure of the virus components. Virology, 37(3), 416–428.
Hull, R., Rees, M. W., & Short, M. N. (1969b). Studies on Alfalfa mosaic virus: I. The protein and nucleic acid. Virology, 37(3), 404–415.
Ibrahim, A., Hutchens, H. M., Berg, R. H., & Loesch-Fries, S. (2012). Alfalfa mosaic virus replicase proteins, P1 and P2, localize to the tonoplast in the presence of virus RNA. Virology, 433(2), 449–461.
Kalinowska, E., Mroczkowska, K., Paduch-Cichal, E., & Chodorska, M. (2014). Genetic variability among coat protein of prune dwarf virus variants from different countries and different Prunus species. European Journal of Plant Pathology, 4, 863–868.
Koonin, E. V., & Dolja, W. (1993). Evolution and taxonomy of positive-strand RNA viruses: Implications of comparative analysis of amino acid sequences. Critical Reviews in Biochemistry and Molecular Biology, 28(5), 375–430.
Korolev, S., Hsieh, J., Gauss, G. H., Lohman, T. M., & Waksman, G. (1997). Major domain swiveling revealed by the crystal structures of complexes of E.Coli rep-helicase bound to single stranded DNA and ADP. Cell, 90, 635–647.
Kozieł, E., Otulak, K., & Garbaczewska, G. (2015). Phylogenetic analysis of PDV movement protein compared to Bromoviridae members as justification of possible intercellular movement. Acta Biologica Cracoviensia Series Botanica, 57(2), 1–9.
Krogh, A., Larsson, B., von Heijne, G., & Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology, 305(3), 567–580.
Liu, L., Westler, W. M., den Boon, G., Wang, X., Diaz, A., Steinberg, H. A., & Alquist, P. (2009). An amphipathic alpha-helix controls multiple roles of Brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathogens, 5(3), 1–18.
Martínez-Gil, L., Sánchez-Navarro, J. A., Cruz, A., Pallas, V., Perez-Gil, J., & Mingarro, I. (2009). Plant virus cell-to-cell movement is not dependent on the transmembrane disposition of its movement protein. Journal of Virology, 83(11), 5535–5543.
Moller, S., Croning, M. D. R., & Apweiler, R. (2001). Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics, 17(7), 646–653.
Olsthoorn, R. C., Mertens, S., Brederode, F. T., & Bol, J. F. (1999). A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO Journal, 18, 4856–4864.
Otulak, K., & Garbaczewska, G. (2010). Ultrastructural events during hypersensitive response of potato cv. Rywal infected withnecrotic strains of potato virus Y. Acta Physiologia Plantarum, 32(4), 635–644.
Otulak, K., Kozieł, E., & Garbaczewska, G. (2016). Ultastructural impact of tobacco rattle virus on tobacco and pepper ovary and anther tissues. Journal of Phytopatology, 164, 226–241.
Paduch-Cichal, E. (2000). Characterization of PNRSV and PDV. Warsaw: Warsaw University of Life Sciences Publishing.
Paduch-Cichal, E., & Sala-Rejczak, K. (2003). Biological properties, stability in crude sap and serogical characterization of prune dwarf virus isolates from Prunus avium seedlings. Phytopathologica Polonica, 29, 9–22.
Pallas, V., Aparicio, F., Herranz, M. C., Amari, K., Sanchez-Pina, M. A., Myrta, A., & Sánchez-Navarro, J. A. (2012). Ilarviruses of Prunus spp.: A continued concern for fruit trees. Phytopathology, 102, 1108–1120.
Pallas, V., Aparicio, F., Herranz, M. C., Sánchez-Navarro, J. A., & Scott, S. W. (2013). The molecular biology of ilarviruses. Advances in Virus Research, 87, 139–183.
Pallas, V., & Garcia, J. A. (2011). How do plant viruses induce disease? Interactions and interference with host components. Journal of General Virology, 92(12), 2691–2705.
PDBViewer program (2016) http://spdbv.vital-it.ch/. Accessed 25.10 2016.
PyMol program (2016) https://www.pymol.org/. Accessed 25.10 2016.
Rafińska, K., Świdziński, M., & Bednarska-Kozakiewicz, E. (2014). Homogalacturonan deesterification during pollen–ovule interaction in Larix decidua mill.: An immunocytochemical study. Planta, 240(1), 195–208.
Ramptish, C., & Estewell, K. C. (1997). The complete nucleotide sequence of prune dwarf ilarvirus RNA-1. Archives of Virology, 142, 1911–1918.
Rozanov, M. N., Koonin, E. V., & Gorbalenya, A. E. (1992). Conservation of the putative methylotransferase domain: A hallmark of the 'Sindbis like' supergroup of positive-strand RNA viruses. Journal of General Virology, 73, 2129–2134.
Sánchez-Navarro, J. A., & Pallas, V. (1997). Evolutionary relationships in the ilarviruses: Nucleotide sequence of Prunus necrotic ringspot virus RNA 3. Archives of Virolology, 142, 749–763.
Schwartz, M., Chen, J., Janda, M., Sullivan, M., den Boon, J., & Ahlquist, P. (2002). A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Molecular Cell, 9, 505–514.
Schwartz, M., Chen, J., Lee, W. M., Janda, M., & Ahlquist, P. (2004). Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. PNAS USA, 101, 11263–11268.
Server AIDA (2016) http://ffas.burnham.org/AIDA/. Accessed 25.10 2016.
Sivakumaran, K., Bao, Y., Roosinck, M. J., & Kao, C. C. (2000). Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of Brome mosaic virus and cucumber mosaic virus. Journal of Virology, 74(22), 10323–10331.
Sivakumaran, K., Chen, M. H., Rosssinck, M. J., & Kao, C. C. (2002). Core promoter for initiation of cucumber mosaic virus subgenomic RNA4A. Molecular Plant Pathology, 3(1), 43–52.
Sonnhammer, E. L. L., von Heijne, G., & Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, 1, 175–182.
Sztuba-Solińska, J., & Bujarski, J. J. (2008). Insights into the single-cell reproduction cycle of members of the family Bromoviridae: Lessons from the use of protoplast systems. Journal of Virology, 82(21), 10330–10340.
Szyndel, M. S., & Paduch-Cichal, E. (1997). Detection and serological comparison of PNRSV and PDV isolates by immunoelectron microscopy techniques. Phytopathologia Polonica, 14, 19–27.
van der Hejden, M. W., Carette, J. E., Reinhoud, P. J., Haegi, A., & Bol, J. F. (2001). Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. Journal of Virology, 75, 1879–1887.
van der Vossen, E. A., Neeleman, L., & Bol, J. F. (1994). Early and late functions of Alfalfa mosaic virus coat protein can be mutated separately. Virology, 202, 891–903.
van der Wel, N. N., Goldbach, R. W., & van Lent, J. (1998). The movement protein and coat protein of Alfalfa mosaic virus accumulate in structurally modified plasmodesmata. Virology, 244, 322–329.
van Pelt-Heerschap, H., Verbeek, H., Huisman, M. J., Loesch-Fries, S., & van Vloten-Doting, L. (1987). Non-structural proteins and RNAs of Alfalfa mosaic virus synthesized in tobacco and cowpea protoplasts. Virology, 161(1), 190–197.
Waterworth, H. E., & Fulton, R. W. (1964). Variation among isolates of Prunus necrotic ringspot virus and prune dwarf virus isolated from sour cherry. Phytopatology, 54, 1155–1160.
Xu, D., Jaroszewski, L., Li, Z., & Godzik, A. (2014). AIDA: Ab initio domain assembly server. Nucleic Acid Research, 42, 308–313.
Xu, D., Jaroszewski, L., Li, Z., & Godzik, A. (2015). AIDA: Ab initio domain assembly for automated multi-domain protein structure prediction and domain-domain interaction prediction. Bioinformatics, 31(13), 2098–2105.
Acknowledgements
The authors would like to thank Tadeusz Malinowski, PhD for providing of PDV isolates inoculum and for his work with DAS-ELISA analyses of leaf samples at Institute of Horticulture in Skierniewice.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kozieł, E., Otulak, K., Lockhart, B.E.L. et al. Subcelullar localization of proteins associated with Prune dwarf virus replication. Eur J Plant Pathol 149, 653–668 (2017). https://doi.org/10.1007/s10658-017-1215-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-017-1215-8