Abstract
Obtaining a sufficient quantity of high-quality, intact RNA is the first crucial step in its study by RNA sequencing on next-generation sequencing platforms or quantitative PCR. Different RNA extraction methods or commercial kits vary in yield and in the quality and integrity of the RNA obtained, which may affect the results of downstream applications. Often, these factors depend on the organism under study and nature of the sample. Therefore, the selection of an appropriate RNA isolation method is critical. In this study, we present the results of an evaluation of three different commercial kits for the isolation of total RNA from Erwinia amylovora in apple tissue as well as the usefulness of different kinds of Deoxyribonuclease I for DNA removal and kits for rRNA depletion. To our knowledge, this is the first report on the method of isolation of high-quality E.amylovora mRNA for RNA-seq.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Erwinia amylovora is a polyphagous bacterium causing fire blight on apple, pear and over 130 other plant species belonging mainly to the Rosaceae family (Van der Zwet and Keil 1979). This pathogen, which causes great economic losses in the areas of apple and pear tree cultivation worldwide, has been the subject of several phenotypic and genetic diversity studies. E. amylovora is generally considered to be a homogeneous species in terms of phenotypic and genetic features (reviewed by Puławska and Sobiczewski 2012). Recent genomic analysis confirmed that the chromosome of E. amylovora is highly conserved and displays over 99 % amino acid sequence identity among all strains tested (Mann et al. 2013). On the other hand, recent application of multi-locus variable number of tandem repeats (MLVA) and clustered regularly interspaced short palindromic repeats (CRISPR) revealed some diversity within E. amylovora (Bühlmann et al. 2014; Rezzonico et al. 2011). In addition, the presence of different plasmids was found to be the source of genetic diversity (McGhee and Jones 2000; Llop et al. 2008; Ismail et al. 2014). Although E. amylovora strains are very homogeneous in terms of phenotypic and genetic features, they show variations in pathogenic ability (Cabrefiga and Montesinos 2005; Hevesi et al. 2000; Puławska et al. 2006; Sholberg et al. 2001; Norelli et al. 1986).
Considering the assumption of, on the one hand, high genomic homogeneity and, on the other hand, high interstrain variation in virulence, it is very interesting to study E. amylovora in terms of the different changes that occur during disease progression following plant infection. In order to study this by RNA sequencing (RNA-seq) based on next-generation sequencing (NGS) platforms or quantitative PCR (qPCR), the first crucial step consists of obtaining a sufficient quantity of high-quality, intact RNA. The first step to the success of an experiment aiming to analyse the bacterial transcriptome in plants includes correct plant inoculation and sampling methods. The next step, the most important - even essential - seems to be the selection of an appropriate RNA extraction method for the study organism. Although laboratory methods have already been described (Mehra 1996), many researchers are using commonly available commercial kits that allow for rapid extraction of high-quality and high-quantity RNA appropriate for high-throughput analysis. However, different RNA extraction kits have proven to vary in yield and level of quality and integrity of the obtained RNA very often, depending on the type of sample (Jahn et al. 2008; Nour et al. 2010; Rump et al. 2010; Deng et al. 2005). Therefore, in order to obtain good quality RNA from the target sample/organism, the different RNA extraction methods or kits should be carefully examined prior to their use in gene expression profiling methods and other applications. Obtaining high values of RNA integrity is critical because low-quality RNA may affect the results of downstream applications, which, in addition to being laborious and time consuming, and are still quite expensive (Nucleic Acids Research 2005).
Independently of the RNA isolation method selected, the researcher must take into account that the majority of kits do not eliminate contaminating genomic DNA, which can adversely affect the results of several applications. Therefore, treatment of the RNA with DNase I, which permits the reduction of DNA content, is a necessary step (Vanecko and Laskowski 1961). For this purpose, different kinds of DNAse I offered by different companies are available.
For the analysis of bacterial mRNA by sequencing, an additional step is necessary. From total RNA, unwanted, abundant rRNA transcripts, constituting approximately 90 % of RNA species, have to be removed. Although the 3′-ends of both prokaryotic and eukaryotic mRNA are polyadenylated, the poly(A) tracts of prokaryotic mRNA are generally shorter, ranging from 15 to 60 adenyl residues and associated with only 2 %–60 % of the molecules of a given mRNA species, which consequently limits the effectiveness of poly(A) enrichment of prokaryotic mRNA species compared with eukaryotic mRNA (Sarkar 1997; Chen and Duan 2011). In the case of bacterial transcriptome analysis, the removal of ribosomal RNA from RNA samples is the best way to prepare mRNA for sequencing; however, not all methods or kits are appropriate for all bacterial species.
The aim of our study was to compare different inoculation methods of apple trees by E. amylovora and different methods of RNA extraction in terms of their ability to obtain high quantity, pure, intact RNA from E. amylovora in plants, as well as to evaluate different DNases for DNA removal and kits for removing rRNA from total RNA.
Materials and methods
Inoculation methods
One-year-old, potted apple trees cv. Idared/M.26 were inoculated with E. amylovora strain 650 in greenhouse conditions in the spring. For inoculations of apple plants, three different methods of inoculation were used: 1) cutting of the tips of the shoots with scissors previously immersed in bacterial water suspension (108 cfu/ml), 2) direct inoculation with a needle: the trees were placed in a horizontal position and shoots were punctured with a sterile needle over approximately 7 cm of their length and covered by 10 μl droplets of bacterial water suspension, approximately 109 cfu/ml prepared by overnight growth in TY medium and 3) direct inoculation as in point 2, but preceded by removal of trichomes from the surface of the shoot. These three different inoculation methods were applied for plants normally watered and for plants that were kept for two days without watering under low humidity conditions before inoculation. After inoculation, the infected plants were covered for 24 h with a plastic bag to maintain high-humidity conditions. Plants were maintained in a quarantine greenhouse at a temperature optimal for symptom development (26 °C) and watered regularly. Control plants were treated in the same way, but instead of bacterial suspension, sterile distilled water was used. RNA isolations were carried out to choose the best method of plant inoculation.
RNA isolations using commercial kits and DNase treatment
After 24 h and six days from the time of inoculation using the selected methods, samples were processed and total RNA was isolated. At each time point, RNA was isolated separately from at least six shoots of each apple cultivar. The inoculated shoots were cut from the plant into pieces and immediately incubated with RNAlater® Stabilization Solution for 25 min with shaking at 26 °C. After that time, the mixture was centrifuged and the pellet washed with sterile distilled water to remove excess RNAlater® Stabilization Solution, which could affect the RNA isolation. For total RNA isolation from the pure culture of E. amylovora strain 650 grown overnight in TY medium and from infected plant material prepared as above, three different commercial kits, each with its own modification, were used: 1) the Promega SV Total RNA System (Promega Corporation, Madison, WI, USA) according to manufacturer’s instructions; 2) the TRIzol® Max™ Bacterial RNA Isolation Kit with two options: with or without the Max™ Bacterial Enhancement Reagent. Some additional modifications were introduced to the methodology: the application of cold or warmed to 60 °C TRIzol reagent and the incubation or not of the collected plant material in RNAlater® Stabilization Solution after the modifications introduced at the beginning. The remaining of the protocol was followed according to the manufacturer’s recommendations, and 3) the Total RNA Purification Kit (Norgen Biotek) according to manufacturer’s instructions.
DNase treatment
As the commercial kits used for RNA isolation in our study did not remove total DNA, which is crucial for downstream applications, two different DNAse treatments were used: DNase I (ThermoScientific) or the TURBO DNA-free™ Kit (Life technologies). The efficiency of removing DNA was tested by nested-PCR with primers Peant1/Peant2 and AJ75/AJ76 (McManus and Jones 1995; Llop et al. 2000) complementary to plasmid pEA29 and primers Ea71/72 (Guilford et al. 1996) complementary to chromosomal DNA. The obtained RNA free from DNA was assessed according to the RNA integrity number (RIN) (Schroeder et al. 2006). Determination of quality and concentration was tested on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) using the Agilent RNA 6000 Nano LabChip® kit according to the manufacturer’s instructions. The RIN algorithm number is from 1 to 10, where level 10 is considered the most intact pure RNA, and 1 represents the most degraded profile of the RNA sample (Mueller et al. 2004).
Depletion of rRNA
As mentioned, to obtain high-quantity and high-quality bacterial mRNA for sequencing, the depletion of rRNA is the crucial step. For this purpose, to remove rRNA from E. amylovora RNA samples, two different kits were tested: the RiboMinus™ Transcriptome Isolation Kit (for bacteria, Lifetechnologies) and the Ribo-Zero™ Magnetic Kit (Gram-Negative Bacteria, http://www.illumina.com).
Results and discussion
The success of all RNA-based analyses depends on the quantity, purity and integrity of the total RNA prepared. An additional requirement for successful bacterial transcriptome analysis by RNA-seq is the efficient removal of ribosomal RNA. However, as presented in this study, there is no universal procedure for all kinds of microorganisms (bacterial species) on all types of plant material or in all other environments, thus it is necessary to select and optimize the protocol for a particular purpose (Nolan et al. 2006). The scientist should have complete control during each step of RNA isolation, taking into account the sensitive nature of RNA, which is easily degraded.
The data described in this study allowed for the selection of the most appropriate inoculation method, a commercial kit for the isolation of high-quality total RNA as well as mRNA free of DNA of E. amylovora for RNA-seq or qPCR analysis. The first step, the selection of the most appropriate apple tree inoculation method showed that, of three different methods of inoculation the third one, involving direct inoculation of shoots with a needle preceded by removal of trichomes from the surface of the shoot, was selected. The removal of trichomes helped in applying droplets of bacterial suspension to the injured plant tissue. It avoided evaporation or loss of the droplets of suspension imposed on the shoot, which resulted in better and faster infiltration of bacteria into plant tissue. In the case of watered plants, the process of penetration of plant tissue by bacterial suspension was 3–4 times longer. The other two methods were less suitable. In the case of the second method, involving direct puncturing with a needle, followed by application of a bacterial suspension, several droplets were lost during inoculation because they easily ran down. Therefore, as repeated inoculation was required, these methods were not applied in further inoculations. In the case of the first method, which has been found to be the best inoculation method, e.g., to test the susceptibility of apple genotypes to fire blight (Sobiczewski et al. 2015) or to compare the virulence of E. amylovora strains (Ismail et al. 2014), no RNA was obtained 24 h after inoculation (data not shown) because of very low bacterial concentrations.
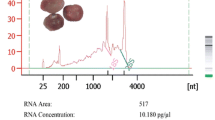
Based on the analysis of total RNA concentration and integrity (RIN parameter), the best quality RNA (RIN 6.5–7.8) was obtained after using the total RNA Purification Kit (Norgen Biotek). In the case of the TRIzol® Max™ Bacterial RNA Isolation Kit, quite good results were obtained only when the TRIzol Reagent was warmed to 60 °C and its use preceded by incubation of the collected plant material in RNAlater® Stabilization Solution: the RIN obtained was approximately 0.5–1 lower from in those samples obtained using the total RNA Purification Kit (Norgen Biotek) (Fig S1). In further analyses, we selected the RNA Purification Kit (Norgen Biotek) as its protocol is less time consuming and avoids the use of harmful chemicals, such as phenol. The worst results in terms of the quality and quantity of isolated RNA were achieved with the Promega SV Total RNA System. Moreover, almost no RNA was isolated in the case of samples processed from plant material with this kit, especially those processed 24 h after inoculation (Fig S2). These RNA samples could not be evaluated by the RIN algorithm and interpreted due to very low RNA concentration obtained and, most likely, also degradation. The minimum RIN recommended for microarray and next generation sequencing analyses is seven, and higher than eight is considered optimal. However, a RIN value higher than five is already considered sufficient for this purpose (Jeffries et al. 2014; Fleige and Pfaffl 2006). The RIN calculation is largely based on the ribosomal RNA ratio, which in the case of E. amylovora, is somehow disturbed because this species possess a 99-bp insertion, representing an intervening sequence – IVS, within the 23S rRNA gene in five of seven copies of the rRNA operon (Smits et al. 2010). IVSs are transcribed but later removed by RNase III without religation during RNA processing, leading to fragmented rRNA (Pronk and Sanderson 2001). Thus, during analysis of RNA concentration and quality we observed four, rather than two, peaks representing rRNAs. Because of this, we could not obtain, in all samples, the ideal RIN value (close to 10); however, in the majority of them, it was possible to obtain a value near the recommended one and a RIN of 7.8 was obtained from the pure bacterial culture (Fig.1). By contrast, with other bacterial genera, eg., Xanthomonas, which possess no IVS in its rRNA and two peaks corresponding to 16S and 23S rRNA, the RIN value obtained with the same kit was near or equal the maximum value of 10 (Fig 2).
Electropherogram of total RNA of Erwinia amylovora strain 650 obtained after isolation with the Total RNA Purification Kit (Norgen Biotek); RNA concentration: 356 ng/μl; RNA Integrity Number (RIN): 6.7. Total RNA was analysed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) using the Agilent RNA 6000 Nano LabChip® kit
Electropherogram of total RNA of Xanthomonas sp. strain obtained after isolation with the Total RNA Purification Kit (Norgen Biotek); RNA concentration: 545 ng/μl; RIN: 10. Total RNA was analysed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) using the Agilent RNA 6000 Nano LabChip® kit
Nowadays, RNA quality is usually assessed by quantification of RNA on ethidium bromide gels (Jakovljevic et al. 2010; Tavares et al. 2011; Sambrook and Russell 2001) or evaluated via measurement of absorbance where an A260/A230 ratio higher than 1.8 is considered as an indicator of extracted RNA with a low level of contamination (Tavares et al. 2011; Pester et al. 2012). In the study presented here, we selected the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) as the most suitable and dedicated method allowing for precise measurements of RNA. Evaluation based on electrophoresis where the sharpness of product visible in the gel is stated as good quality (Jakovljevic et al. 2010) seems imprecise, as it relies on the human interpretation of a gel image (Schroeder et al. 2006), but spectrometric methods are not very sensitive and do not give an answer about RNA integrity.
Of the two DNases used for the removal of genomic DNA, DNase I (ThermoScientific) turned out to be more efficient. Based on PCR amplification with specific primer pairs, a lower number of DNase treatments was necessary in comparison with the TURBO DNA-free™ Kit (Life technologies), usually 1–2 compared with 3–5, respectively. The level of DNA contamination may be genus/species and RNA isolation method specific. Generally 1–2 treatments of RNA sample with DNase is considered as the necessary minimum (Jahn et al. 2008). From the two different primer pairs tested, the primers Peant1/Peant2 and AJ75/AJ76 were much more sensitive and allowed us to detect even small amounts of DNA in the samples where it was not detected using the primers Ea71/72. This is related to the fact that these first two primer pairs are complementary to plasmid pEA29, which occurs in one to five copies per bacterial cell (Mann et al. 2013; Smits et al. 2010), while primers Ea71/72 are complementary to chromosomal DNA.
Out of two kits used for removal of rRNA, the Ribo-Zero™ Magnetic Kit*(Gram-Negative Bacteria) was selected for processing of all samples. Using the RiboMinus™ Transcriptome Isolation Kit (for bacteria), the 5S rRNA fraction was not removed, but 16S and 23S rRNA were only partially removed (Fig S3). The removal of 5S rRNA was not achieved because this kit is not predicted to remove this fraction of rRNA, while 23S rRNA could not be completely removed due to fragmentation. Using the Ribo-Zero™ Magnetic Kit*(Gram-Negative Bacteria), it was possible to remove from total RNA all of the rRNA particles (Fig S3). This is probably connected with an oligonucleotide probe mixture in the kit containing two probes targeting 16S rRNA and three probes targeting 23S rRNA (Chen and Duan 2011 ), which turned out to be compatible with all rRNA particles of E. amylovora. On the market there are other kits for bacterial rRNA removal like MICROBExpress Kit (Ambion) and mRNA-ONLY™ Prokaryotic mRNA Isolation Kit (Epicentre), however based on the results of other authors we knew that they were not able to fully remove rRNA, particularly when rRNA subunits are not intact as in case of E. amylovora (Ciulla et al. 2010) so we did not include them in our study.
Thanks to the study presented here, we can recommend for other researchers the appropriate kits that are useful for total RNA and mRNA of E. amylovora extraction for RNA-seq or gene expression analyses.
References
Bühlmann, A., Dreo, T., Rezzonico, F., Pothier, J. F., Smits, T. H. M., Ravnikar, M., et al. (2014). Phylogeography and population structure of the biologically invasive phytopathogen Erwinia amylovora inferred using MLVA. Environmental Microbiology, 16(7), 2112–2125.
Cabrefiga, J., & Montesinos, E. (2005). Analysis of aggressiveness of Erwinia amylovora using disease-dose and time relationships. Phythopathlogy, 95, 1430–1437.
Chen, Z., & Duan, X. (2011). Ribosomal RNA depletion for massively parallel bacterial RNA-sequencing applications. Methods in Molecular Biology, 733, 93–103.
Ciulla, D., Giannoukos, G., Earl, A., Feldgarden, M., Gevers, D., Levin, J., et al. (2010). Evaluation of bacterial ribosomal RNA (rRNA) depletion methods for sequencing microbial community transcriptomes. Genome Biology, 11(Suppl 1), P9.
Deng, M. Y., Wang, H., Ward, G. B., Beckham, T. R., & McKenna, T. S. (2005). Comparison of six RNA extraction methods for the detection of classical swine fever virus by real-time and conventional reverse transcriptase-PCR. Journal of Veterinary Diagnostic Investigation, 17, 574–578.
Fleige, S., & Pfaffl, M. W. (2006). RNA integrity and the effect on the real-time qRT-PCR performance. Molecular Aspects of Medicine, 27, 126–139.
Guilford, P. J., Taylor, R. K., Clark, R. G., Hale, C. N., Forster, R. L. S., & Bonn, W. G. (1996). PCR-based techniques for the detection of Erwinia amylovora. Acta Horticulture, 411, 53–56.
Hevesi, M., Papp, J., Jambor-Benczur, E., Kaszane-Csizmar, K., Pozsgai, I., Gazdag, G., et al. (2000). Testing the virulence of some Hungarian Erwinia amylovora strains on in vitro cultured apple rootstocks. International Journal of Horticultural Science, 6, 2–55.
Ismail, E., Blom, J., Bultreys, A., Ivanovic, M., Obradovic, A., van Doorn, J., et al. (2014). A novel plasmid pEA68 of Erwinia amylovora and the description of a new family of plasmids. Archives of Microbiology, 196, 891–899.
Jahn, C. E., Charkowski, A. O., & Willis, D. K. (2008). Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. Journal of Microbiological Methods, 75, 318–324.
Jakovljevic, K. V., Spasic, M. R., Malisic, E. J., Dobricic, J. D., Krivokuca, A. M., & Jankovic, R. N. (2010). Comparison of phenol-based and alternative RNA isolation methods for gene expression analyses. Journal of Serbian Chemical Society, 75, 1053–1061.
Jeffries, M. K. S., Kiss, A. J., Smith, A. W., & Oris, J. T. (2014). A comparison of commercially-available automated and manual extraction kits for isolation of total RNA from small tissue samples. BMC Biotechnology, 14, 94.
Llop, P., Bonaterra, A., Penalver, J., & Lopez, M. M. (2000). Development of a highly sensitive nested-PCR procedure using single closed tube for detection of Erwinia amylovora in asymptomatic plant material. Applied and Environmental Microbiology, 66, 2071–2078.
Llop, P., Gonzalez, R., Pulawska, J., Bultreys, A., Dreo, T., & Lopez, M. M. (2008). The new plasmid pEI70 is present in Erwinia amylovora European strains. Acta Horticulture, 793, 131–136.
Mann, R. A., Smits, T. H. M., Buhlmann, A., Blom, J., Goesmann, A., Frey, J. E., et al. (2013). Comparative genomics of 12 strains of Erwinia amylovora identifies a pan-genome with a large conserved core. PloS One, 8(2), e55644.
McGhee, G. C., & Jones, A. L. (2000). Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and interspecies variation. Applied and Environmental Microbiology, 66, 897–4907.
McManus, P. S., & Jones, A. L. (1995). Detection of Erwinia amylovora by nested PCR, PCR-dot-blot and reverse-blot hybridisation. Phytopathology, 85, 618–623.
Mehra, M. (1996). RNA isolation from cells and tissues. In P. Krieg (Ed.), A laboratory guide to RNA: isolation, analysis and synthesis (pp. 1–20). N.Y.: Wiley-Liss.
Mueller, O., Lightfoot, S., Schroeder, A., 2004. RNA Integrity Number (RIN)—Standardization of RNA Quality Control. Agilent Technologies, Technical Report, 5989-1165EN, 1–8.
Nolan, T., Hands, R. E., & Bustin, S. A. (2006). Quantification of mRNA using real-time RTPCR. Nature Protocols, 1, 1559–1582.
Norelli, J. L., Aldwinckle, H. S., & Beer, S. V. (1986). Differential susceptibility of malus spp. Robusta 5, Novole, and Ottawa 523 to infection by Erwinia amylovora. Plant Disease, 70, 1017–1019.
Nour, A. M., Barbour, E. K., Depint, F., Dooms, M., Niang, K., Dulac, A., et al. (2010). Comparison of five RNA extraction methods from rabbit’s blood. Agriculture and Biology Journal of North America, 1, 448–450.
Imbeaud, S., Graudens, E., Boulanger, V., Barlet, X., Zaborski, P., Eveno, E., Mueller, O., Schroeder, A., Auffray, C. 2005., Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces Nucleic Acids Research, 33, 1–12.
Pester, D., Milcevicova, R., Schaffer, J., Wilhelm, E., & Blumel, S. (2012). Erwinia amylovora expresses fast and simultaneously hrp/dsp virulence genes during flower infection on apple trees. PloS One, 7(3), e32583.
Pronk, L. M., & Sanderson, K. E. (2001). Intervening sequences in rrl genes and fragmentation of 23S rRNA in genera of the family Enterobacteriaceae. Journal of Bacteriology, 183, 5782–5787.
Puławska, J., & Sobiczewski, P. (2012). Phenotypic and genetic diversity of Erwinia amylovora: the causal agent of fire blight. Trees – Structure and Function, 26, 3–12.
Puławska, J., Kielak, K., & Sobiczewski, P. (2006). Phenotypic and genetic diversity of selected polish Erwinia amylovora strains. Acta Horticulture, 704, 439–444.
Rezzonico, F., Smits, T. H. M., & Duffy, B. (2011). Diversity, evolution and functionality of clustered regularly interspaced short palindromic repeat (CRISPR) regions in the fire blight pathogen Erwinia amylovora. Applied and Environmental Microbiology, 77, 3819–3829.
Rump, L. V., Asamoah, B., & Gonzalez-Escalona, N. (2010). Comparison of commercial RNA extraction kits for preparation of DNA-free total RNA from salmonella cells. BMC Research Notes, 3, 211.
Sambrook, J., & Russell, D. W. (2001). Molecular cloning. A Laboratory Manual: Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
Sarkar, N. (1997). Polyadenylation of mRNA in prokaryotes. Annual Review of Biochemistry, 66, 173–197.
Schroeder, A., Mueller, O., Stocker, S., Salowsky, R., Leiber, M., Gassmann, M., et al. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology, 7, 3.
Sholberg, P. L., Bedford, K. E., Haag, P., & Randall, P. (2001). Survey of Erwinia amylovora isolates from British Columbia for resistance to bactericides and its virulence on apple. Canadian Journal of Plant Pathology, 23, 60–67.
Smits, T. H. M., Rezzonico, F., Kamber, T., Blom, J., Goesmann, A., Frey, J. E., et al. (2010). Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Molecular Plant-Microbe Interactions, 23, 384–393.
Sobiczewski, P., Peil, A., Mikicinski, A., Richter, K., Lewandowski, M., Zurawicz, E., et al. (2015). Susceptibility of apple genotypes from European genetic resources to fire blight (Erwinia amylovora). European Journal of Plant Pathology, 141, 51–62.
Tavares, L., Alves, P. M., Ferreira, R. B., & Santos, C. N. (2011). Comparison of different methods for DNA-free RNA isolation from SK-N-MC neuroblastoma. BMC Research Notes, 4, 3.
Van der Zwet, T., & Keil, H. L. (1979). Fire blight—a bacterial disease of rosaceous plants. In Agricultural handbook 510. U.S.: Department of Agriculture, Washington DC.
Vanecko, S., & Laskowski, M. S. (1961). Studies of the specificity of deoxyribonuclease I. II. Hydrolysis of oligonucleotides carrying a monoesterified phosphate on carbon 3′. Journal of Biological Chemistry, 236, 1135–1140.
Acknowledgments
This work was financed by the National Science Centre, Poland, Grant UMO-2012/05/B/NZ9/03455. The authors would like to thank Mrs. Halina Kijańska for excellent technical help.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(PDF 545 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kałużna, M., Kuras, A., Mikiciński, A. et al. Evaluation of different RNA extraction methods for high-quality total RNA and mRNA from Erwinia amylovora in planta . Eur J Plant Pathol 146, 893–899 (2016). https://doi.org/10.1007/s10658-016-0967-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-016-0967-x