Abstract
Tomato fruits can be contaminated by saprophytic strains of Alternaria alternata which is the reason for the frequent occurrence of Alternaria toxins like alternariol, alternariol monomethylether or tenuazonic acid in these types of products. It was shown earlier that alternariol is a colonization factor for tomatoes. In the current analysis two different tomato genotypes were analysed by untargeted comprehensive two-dimensional gas chromatography mass spectrometry (GC×GC-MS). This analysis revealed clear differences in the metabolic profiles which were paralleled by differences in resistance towards Alternaria colonization. One of the genotypes was more resistant against A. alternata infection and contained high amounts of chlorogenic acid in contrast to the other genotype which was sensitive against infection. In in vitro analysis, chlorogenic acid reduced alternariol biosynthesis during the first days of growth of A. alternata. Expression analysis of the alternariol polyketide synthase gene, a key gene in the biosynthesis of alternariol, also revealed a temporal reduction in its expression in the first phases of growth. However by chromatographic analysis it could be demonstrated that chlorogenic acid was degraded over time. This degradation leads to a relief of inhibition resulting in an only temporal inhibition of alternariol biosynthesis. In vivo colonization experiments revealed that chlorogenic acid reduces colonization of tomatoes by A. alternata in a concentration dependent manner, which however is partly counteracted by the addition of alterariol.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A. alternata is an important plant pathogenic and toxigenic fungus for certain plants. Pathogenic strains of this species are responsible for various plant diseases such as black and grey rot of citrus fruit, black point of cereals, black rot of carrots, sunflower, pepper, melon and black rot of tomatoes (Logrieco et al. 2009). Pathogenic strains of A. alternata can directly infect the plant in the field and typically lead to the described rot-like plant pathogenic processes. All of these strains carry host specific toxins like the AAL toxin for the tomato specific infection. These toxins typically lead to an induction of necrosis and suppression of host defence mechanisms to facilitate the infection process (Akamatsu et al. 1997). In contrast to the pathogenic strains, saprophytic strains do not produce these host specific toxins and therefore have a lower capacity for host colonization. These strains usually infect the fruit during post-harvest at a high water activity and need lesions due to sunburn or bruising of the ripe fruit. Morris et al. (2000) showed that from 62 strains of A. alternata isolated from Californian tomatoes, only one strain was able to infect tomato leaves suggesting that most strains isolated from tomato fruits were non-pathogenic. In another analysis (Graf 2012) a very similar result was described. From 63 strains of A. alternata isolated from tomato fruits, none was pathogenic but all were saprophytic. This was determined by a specific PCR reaction directed against the polyketide synthase gene which is involved in the biosynthesis of AAL toxin (Graf 2012). Despite the low pathogenic activity of these Alternaria strains, tomatoes and tomato products are very frequently contaminated with Alternaria toxins like alternariol (AOH), alternariol monomethylether (AME) or tenuazonic acid (TeA), sometimes even in very high amounts (Ostry 2008; Ackermann et al. 2012). Interestingly all of the 63 strains mentioned above were high and consistent AOH producers. In view of this it was recently shown that AOH is a colonization factor for saprophytic infection of tomatoes (Graf 2012). Mutants of A. alternata, which were no longer able to produce AOH, were not able to colonize wounded tomato tissue, whereas the wild type could. The addition of purified AOH to the infection side increased the colonization rate of A. alternata significantly. So the consistent production of AOH by saprophytic strains of A. alternata supports efficient colonization of tomatoes.
On the other hand, even wounded tomatoes are not completely defenceless against fungal infection. Tomatoes produce various secondary metabolites with antifungal activity. One very well-known metabolite is tomatin, a saponin found in various fruits, but especially tomatoes (Osbourn 1996; Arie et al. 2007). Saponines form complexes with the sterols of fungal membranes which lead to the formation of pores and to a loss of cell wall integrity. However, the protection of tomatine towards certain fungi is only transient, because several fungi like Septoria lycopersici, Verticillium albo-atrum, Fusarium oxysorum or A. alternata can degrade tomatin by the aid of a tomatinase to non-toxic degradation products (Osbourn 1996). A further important fungus defence compound is chlorogenic acid (CGA). CGA is a polyphenol. Structurally it is an ester between caffeic acid and L-quinic acid and occurs in quite high amounts in solanaceous plants like potatoes and tomatoes (Ahmad et al. 2013). It has been described several times as an important antifungal secondary metabolite from tomatoes (Ruelas et al. 2006; Atanasova-Penichon et al. 2012). Ruelas et al. (2006) analysed the polyphenolic acid composition of tomatoes and found that caffeic, p-coumaric, ferulic and CGA, were the most important polyphenols in various varieties of tomatoes. CGA has important concentration dependent antifungal activities. Bostock et al. (1999) showed that the resistance of peach against Monilia fructicola decreased with the reduction of CGA content. This could also be confirmed by Terry et al. (2014) who showed that depending on decreasing concentrations of phenolic acids, strawberries were more susceptible towards infection by Botrytis cinerea. So based on this data, CGA has clearly antifungal properties and protects tomatoes from fungal attack.
The inherent and preformed plant defence mechanisms, like for example the presence of polyphenols and other compounds, inhibit the attacking fungus, but the fungus reacts with the activation of various pathogenicity and colonization factors. As described above, the biosynthesis of AOH is such a reaction. AOH is a mycotoxin, which, e. g. beside its role in the colonization of tomatoes (Graf 2012) exhibits toxic activities against humans and animals (Ostry 2008). Other mycotoxins which also constitute colonization factors are patulin, produced by Penicillium expansum, the main pathogenic organism of apples (Sanzani et al. 2012) and the trichothecenes, produced among other mycotoxins by F. graminearum (Desjardins et al. 1992). The importance of alternariol for a successful colonization of tomatoes denotes that a reduction in AOH biosynthesis would reduce the colonization capacity by A. alternata.
AOH, or in its methylated form AME, are polyketide mycotoxins, meaning that in their biosynthesis a polyketide synthase is involved. Polyketide synthases are usually large multifunctional enzymes, which are coded by one gene, which usually is located within a cluster of biosynthesis genes of the respective mycotoxin. Recently the pksJ gene has been identified (Saha et al. 2012) which is responsible for the biosynthesis of AOH and AME. This enables the analysis of the expression of this gene, in relation to environmental conditions.
Untargeted metabolomics is a powerful technique to identify and quantify a large number of primary and secondary metabolites in tomatoes or other plants (Moco et al. 2006; Osorio et al. 2012). Compared to LC/MS and one-dimensional GC/MS, GC×GC-MS combines a very high separation power, a high sensitivity and the possibility to identify unknown metabolites using dedicated EI mass spectral libraries (Mondello et al. 2008). This kind of analysis is therefore suitable to compare different tomato genotypes and to identify metabolites specific for each genotype.
In the current analysis, the metabolome of two tomato varieties, one resistant and one susceptible to the infection by A. alternata, were analysed by GC×GC-MS. Among other metabolites, CGA was one of the most discriminating metabolite with respect to fungal resistance. The influence of this compound on growth, expression of the pksAL gene and thereby on AOH biosynthesis was analysed. It could be shown that CGA transiently inhibits AOH biosynthesis. CGA is however degraded after prolonged incubation. In a subsequent in vivo colonization experiment the results obtained in vitro could be confirmed.
Materials and methods
Fungal strains and growth conditions
A. alternata BFE1316, originally isolated from ripe tomato fruits, is a consistent AOH producing strain and was used as a model strain throughout this study. For strain maintenance, strain propagation and for the experiments with CGA, the strain was grown on MG medium (malt extract medium, Merck, Darmstadt, Germany; supplemented with 5 g/l glucose). Tomato medium was used as a model medium for the natural environment of A. alternata. For the preparation of tomato medium, 400 g of commercially available tomatoes were minced and mixed together with 15 g agar with bi-distilled water to get a final volume of 1 l, which then was autoclaved.
Determination of AOH and AME by TLC and HPLC
After 7 days of incubation at 25 °C, two agar plugs (diameter 8 mm) with adhering mycelium were isolated with a corer from the colony, placed in a micro reaction tube and extracted with 1 ml chloroform under shaking conditions for 30 min. The mycelial debris was removed, the dissolvent was evaporated in a Savant SpeedVac Concentrator (Thermo Fisher Scientific, Waltham, USA) and the residue resolved in 100 μl methanol. An amount of 30 μl of each mycotoxin extract was applied on a TLC F254 silica plate (Silica gel 60, Merck, Darmstadt, Germany) and developed with toluol: ethylacetate:formic acid (60:35:5) as mobile phase. AOH (Alexis Biochemicals, Lörrach, D, 0.2 ng/ml in methanol) was used as standard. The air dried plates were visualized and photographed at 365 nm for 700 ms (TLC Visualizer, Camag, Muttenz, CH) and quantified at 337 nm (TLC Scanner, Camag, Muttenz, CH) if required.
HPLC was performed in a Hitachi system, consisting of a D7100 pump, a D7000 sample applicator, a L7360 column oven and a L7455 diode array detector. A Nucleodur C18 Pyramid 3 pre- and main column (Machery and Nagel, Germany) were used for separation. A volume of 10 μl was applied onto the HPLC and separated at a temperature of 40 °C at a flow rate of 0.5 ml/min. As an effluent, a gradient between methanol/427.5 nM phosphoric acid from 40/60 to 95/5 was used. The detection was performed with the diode array detector.
Sampling and sample preparation of tomato fruit tissue
The tomato varieties Schmucktomate (ST) and Resi were obtained from the Organic Outdoor Tomato Project (Horneburg and Becker, 2011) of the University of Göttingen, Germany. Ripe fruits from four field replicates per genotype, consisting of two plants each, were harvested, transported to the laboratory in shock-protected boxes and stored overnight at 10 °C. One pooled sample was prepared for each field replicate combining 6–10 fruits, depending on fruit size. From each fruit, one quarter was cut out longitudinally with a scalpel. The exocarp, the placenta and the seeds were discarded. The remaining parts of the pericarp were chopped, frozen in liquid nitrogen and freeze-dried for 48 h. The dried tissue pieces were ground using a ball mill (Retsch MM200) for 1 min at 25 Hz. For each sample, two powder aliquots of 20 ± 0.06 mg, for duplicate analysis, were weighed out in 2 ml Eppendorf reaction tubes. Further powder aliquots from each genotype were combined to prepare a representative pooled quality control (QC) sample from which 21 QC aliquots were prepared subsequently in the same manner as the study samples. All powder aliquots were stored at −80 °C until analysis.
Extraction and derivatization of freeze-dried tomato samples
On the day of analysis, the samples were thawed on ice. After the addition of 750 μl of methanol to the powder, the samples were spiked with 30 μl of a solution containing eight internal standards (d-pinitol, ribitol, 1-o-methyl-2-desoxy-d-ribose, 5-bromo-2,4-dihydroxybenzoic acid, 5-chlorosalizylic acid, pentadecanoic acid, n-methyl-l-serine and 2-(4-chlorophenyl)ethylamine, each 2.5 or 5 mM in 62,5 % EtOH) and mixed for 10 min at 30 °C and 1,400 rpm. The insoluble matter was sedimented by a short centrifugation and 700 μl of the supernatant were transferred to a new Eppendorf reaction tube. The samples were re-extracted a second time, both supernatants were combined and finally centrifuged for 5 min at 16,100 × g and 4 °C. 20 μl of the supernatant were transferred to screw-threaded GC vials containing 200 μl inserts and evaporated in a vacuum centrifuge for 1.5 h at 40 °C and approx. 1 mbar. For methoximation, 30 μl of methoxylamine-hydrochloride in dried pyridine (20 mg/ml) were added and samples incubated for 1 h at 40 °C under shaking. Trimethylsilylation was initiated by the addition of 60 μl MSTFA +1 % TMCS and carried out for 1 h at 75 °C without shaking. An amount of 8 μl of a linear retention index mixture (saturated fatty acid methyl esters (C7 to C26), each 250 μM in heptane) were added only to daily reagent controls after derivatization. All samples were analysed within 24–30 h after preparation.
GC×GC-MS analysis
The GC×GC-MS system consisted of a Shimadzu GCMS QP2010 Ultra instrument, equipped with an AOC-5000 autosampler (Shimadzu, Duisburg, Germany), an OPTIC-4 PTV injection system (ATAS GL International B.V., Eindhoven, The Netherlands) and a ZX2 cryogenic dual-jet, air-based loop modulator (ZOEX Corp., Houston, USA). The first column (Restek Rxi-5SilMS, 15 m × 0.25 mm × 0.25 μm) was connected to the second column (SBE BPX50, 2.5 m × 0.15 mm × 0.15 μm; 1.4 m for modulator loop and 1.1 m for separation) using a SilTite MiniUnion connector (SGE). The initial temperature of the PTV injector was 70 °C; after injection of a sample volume of 1 μl in split mode (1:10), the temperature was raised immediately to 280 °C at a rate of 60 °C/min and held until the end of the run. GCxGC separation was achieved using the following temperature program: 90 °C – 2 °C/min – 98 °C – 6 °C/min – 140 °C – 3 °C/min – 200 °C – 4 °C/min – 245 °C – 3 °C/min – 270 °C – 60 °C/min – 330 °C (3.42 min) and took 55 min per run. The GC was operated in constant linear velocity mode (32.8 cm/s). The initial hot–jet temperature of the modulator (200 °C) was changed according to the following program: 250 °C (t = 10 min) – 330 °C (t = 30 min) – 200 °C (t = 54 min). We choose a modulation period of 4.5 s and a hot-jet pulse duration of 375 ms. The quadrupol MS (qMS) detector was operated at maximum scan speed (20,000 u/s), with an event time of 30 ms. EI spectra (70 eV) were recorded in scan mode, covering a range of m/z 60–550. Data acquisition began after a solvent cut of 3.5 min. The temperatures of the MS interface and the ion source were 330 and 200 °C, respectively. The GC×GC-MS instrument was controlled using GCMS Solution (version 2.72); the OPTC-4 PTC injector was controlled using the Evolution Workstation software.
A number of 64 study samples, 17 QC samples and four solvent controls were analysed in four batches on four consecutive days; because every QC sample was injected twice, a total number of 102 runs resulted. A general daily run order scheme was developed which started with the daily reagent control, followed by three blocks of five study samples which where each enclosed by twin injections of QC samples. Thus, the reagent control, 15 study samples, as well as four QC samples (two runs each), were analysed per day. On the last day, a fourth block consisting of four study samples and an additional QC sample was attached to complete the measurement.
Data processing and statistical analysis
Data processing comprised two main steps. At first, all peaks were picked and integrated with the Postrun Analysis feature of GCMS Solution. Because of the high number of raw data peaks, several partially overlapping chromatogram segments were integrated by an automated batch processing procedure. The areas and heights of the total ion current (TIC) peaks were determined (slope =5,000 min−1; width =0.04 s; drift =1,000 min−1; T.DBL =1,000 min; minimal area =10,000; no smoothing) and peak apex spectra were saved after baseline subtraction. For compound identification, the spectra were matched against a mass spectral library developed in-house, including linear retention indices (LRI) based on saturated fatty acid methyl esters. Peaks were annotated, if spectral similarity was at least 75 % and the LRI deviation less than +15 or −10 units. Finally, the results (detected peaks, library search results and mass spectra) were exported as one text file per sample. For data visualization purposes, the raw data was converted to the netCDF format.
All further steps, such as peak preprocessing, peak alignment of all sample runs and statistical analysis, were performed with in-house developed r- modules (v. 3.0.1; R Core Team 2013, A language and environment for statistical computing.(www.R-project.org). Hypothesis testing (volcano plot) was calculated with an unpaired, two-sided t-test, assuming unequal variances and a confidence level of 95 %. For all other data the average of triplicate values and the SD values are given.
Isolation of RNA
To perform gene expression experiments, RNA was isolated using the RNAeasy Plant Mini kit (Qiagen, Hilden, Germany). An amount of 1.0 g of the mycelium was ground in a mortar in the presence of liquid nitrogen. For normalization of the subsequent Real Time PCR, an amount of exactly 250 mg of the mycelium powder, determined by a microbalance, was used for isolation of total RNA. RNA concentration was determined by a Nanodrop spectrophotometer (Peqlab, Erlangen, Germany). The mycelium was resuspended in 750 ml lysis buffer, mixed with 7.5 ml β-mercaptoethanol and about 100 glass beads with a diameter of 1 mm (B. Braun Biotech International GmbH, Melsungen, Germany) in a RNase free micro reaction tube. The sample was mixed thoroughly and incubated for 15 min at 55 °C and 42 kHz in an S10H ultrasonic bath (Elma, Singen, Germany). All further procedures were essentially the same as suggested by the manufacturer of the RNA isolation kit.
cDNA synthesis
For cDNA synthesis, 12 ml of the DNase I treated total RNA were used along with the Omniscript Reverse Transcription kit (Qiagen, Hilden, Germany). The reaction mixture was composed essentially as described by the manufacturer and incubated at 37 °C for 1 h. The cDNA was either directly used for Real Time PCR or stored at –20 °C.
Real Time PCR for expression analysis of the pksJ gene
The Real Time PCR reactions were performed in an IQ5 Real Time PCR system (BIO-Rad, Munich, Germany). The SYBR Green approach was used. The primers were developed according to the published sequence of pksJ, the alternariol polyketide synthase gene decribed by Saha et al. (2012; GeneBank accession number: JX103645.1). The primer set had the following nucleotide sequences: Alta-pks1_SY_for (5- ’gat ctc tca gcg tga ttc tca g-3’) and Alta_pks1_SY_rev (5’- ccg tat caa ccg tca tgc t-3’). For the PCR reaction, the SYBR Green Reagent kit (BioRad) was used according to the recommendations of the manufacturer. After an activation step of 10 min at 95 °C, all subsequent 40 PCR cycles were performed according to the following temperature regime: 95 for 20, 55 for 40, and 72 °C for 60 s. A standard curve was generated for absolute quantification of the pksJ mRNA copies. To generate this standard curve, a larger PCR fragment of the pksJ gene was used as template, which was amplified with the primer pair Alta_pks1_for (5’-ggt ttc tac aaa gag cat ggc-3’) and Alta_pks1_rev (5’-gtc gct ctt gag cag att gg-3’). The concentration of this standard PCR product was determined in a Nanodrop spectrophotometer and the number of copies was calculated. These stock solutions were diluted serially by a factor of ten and an aliquot of the dilutions was used as a copy number standard during each setup of the Real Time PCR reaction.
Colonization studies on tomatoes
For the colonization experiments with tomatoes from an A. alternaria BFE1316 culture grown for 7 days on MEA agar at 20 °C, a spore suspension of a concentration of 103 spores/ml in physiological NaCl solution (0.75 % NaCl) was prepared. This solution either was used directly for infection of tomatoes, or a CGA or AOH solution was added to reach a concentration of 65 or 650 ng/ml, in the case of CGA and, or of 0.1 μg/ml, in the case of AOH. These solutions were prepared in such a way, that the final spore concentrations were 103 spores/ml, too. For the application of the spore suspensions, the tomatoes (commercially available) were wounded with a sterile scalpel by cutting 5 mm into the cuticula of the tomato. A volume of 10 μl of the spore suspension was applied onto this wound with a microliter pipet. The infected tomatoes were placed into glass beakers and incubated for up to 7 days at 20 °C. After that time, the tomatoes were visually inspected and 1 g of tomato tissue around the fungal colony was isolated by a sterile scalpel and extracted by chloroform for HPLC analysis.
Results
Comparison of the colonization efficiency and AOH biosynthesis by an A. alternata strain on different tomato genotypes
In order to analyse the colonization efficiency of A. alternata BFE1316 and its AOH biosynthesis capacity on different tomato genotypes, a spore suspension of a 5 day old colony of A. alternata BFE1316 was prepared. The spore suspension was adjusted to 103 spores/ml. A volume of 10 μl of this spore suspension was used to inoculate at least 8 wounded tomato fruits per genotype. The fruits were incubated 20 °C for 7 days. The tomato genotypes were Resi and ST. After an incubation of 7 days, clearly a different colonization behaviour of A. alternata BFE1316 became obvious. On genotype ST A. alternata colonized single fruits only very sparsely. In contrast to this, a high colonization and growth of A. alternata BFE1316 could be observed on Resi. These results indicate that ST has a much more resistant phenotype than Resi. To analyse the AOH produced under these conditions, 1.5 g of tomato tissue from below the fungal colony was isolated and subjected to HPLC. The results are shown in Fig. 1. In congruence with the resistance against colonization, also the amount of AOH found in the tomato genotypes is higher in the sensitive Resi genotype compared to the resistant ST genotype. So there was a clear tendency that the genotype Resi contained more AOH than the genotype ST.
Taken together this is an argument that the presence of high concentrations of AOH favors colonization.
Identification of differentially produced metabolites in the two tomato genotypes by untargeted GC×GC-MS
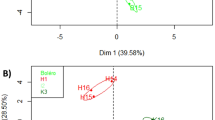
By using the untargeted GC×GC-MS approach described above 267 metabolites could be consistently detected and reproducibly quantified in tomato samples by means of relative quantification. To identify metabolites most responsible for the differences between genotypes ST and Resi and to visualize the respective fold changes a volcano plot (Fig. 2) was used. The metabolites which showed the highest and statistically significant differences (p < 0.05) between the two genotypes are given in Table 1. CGA proved to be the most discriminating metabolite in the more resistant ST genotype. Additionally, besides several unknown compounds, primary metabolites like putrescine, ethanolamine and the amino acids glutamine and leucine, were also elevated. The dehydroascorbic acid derivatives might result from oxidation of ascorbic acid during derivatization. Nicotinic acid was present only in traces. On the contrary, higher levels of several unknown sugar-like compounds, as well as organic acids (saccharic, galacturonic, glucuronic, gluconic, succinic, 2-hydroxyglutaric, quinic and threonic acid), were characteristic for the tomato genotype Resi. Because antifungal effects have already been described for CGA (Ruelas et al., 2006; Suárez-Quiroz et al., 2013), we chose this compound for further investigation.
Volcano plot of the metabolite profile of two the tomato varieties ST and Resi. ST is a resistant variety against A. alternata, whereas Resi is sensitive. The secondary metabolite (CGA) which occurs in high concentrations in the ST variety is highlighted. The concentration of the metabolites below the green lines (yellow area) are not significantly different between the two tomato varieties, whereas the concentrations of the metabolites in the blue areas show an at least two fold change and are thus most discriminating. The metabolites on the left side (left blue area) are higher in the tomato variety ST2, the metabolites on the right side (right blue area) are higher in the tomato variety Resi
Influence of chlorogenic acid on AOH, AME und TeA biosynthesis
Because it was recently shown that AOH is a colonization factor of A. alternata for the infection of tomatoes, the influence of CGA on growth and the biosynthesis of AOH, but also AME and TeA by A. alternata BFE1316 were analysed. For that purpose, A. alternata BFE1316 was inoculated on MEA medium with increasing concentrations of CGA (0 – 3 mg/ml) and grown for up to 7 days at 20 °C. After 3, 4 and 7 days the growth capability of A. alternata was determined and the AOH, AME and TeA produced was analysed by HPLC. There was also a CGA concentration and time dependent reduction of AOH and AME biosynthesis. Strongest AOH and AME biosynthesis was on MEA medium without added CGA (Fig. 3). Under these conditions already at day 4 reasonable amounts of both metabolites were produced. However almost no AOH or AME could be detected at all concentrations of CGA at day 4. If however incubation proceeded to day 7 AOH and AME could be produced, but to a strongly reduced rate compared to the control. At the highest concentration (3 mg/ml) very low amounts of AOH and AME were produced even at day 7. This behaviour indicates that either A. alternata adapts to the activity of CGA or more likely that CGA is degraded over time.
In contrast to the inhibition of AOH and AME, TeA seems to be induced by CGA (Fig. 3c). At 1 mg/ml the TeA concentration was increased compared to the control and was the highest under the conditions analysed. A further increased of CGA to 3 mg/ml than reduced the amount of TeA below the concentration of the former condition. Beside this effect the production kinetics of TeA differed from that of AOH/AME. The latter two metabolites showed a “typical” production kinetics of a secondary metabolite, starting with low amounts at the early times of growth and reaching high amounts at later times. Interestingly in the case of TeA high amounts of the metabolite are produced over the whole observation range indicating a less regulated production of this metabolite.
Taken together the results show that CGA specifically inhibits AOH and AME biosynthesis in A. alternata in a concentration dependent manner. However, according to the results shown, this inhibition is transient, being partly relieved after prolonged incubation times. This could either be due to an adaptation of A. alternata to the inhibiting effect of CGA or by a degradation of CGA.
Degradation of CGA
During the cause of the experiments it became obvious, that in the chromatograms of the AOH non-producing cultures on CGA medium a new signal could be detected, which disappeared during the shift to AOH producing conditions (Fig. 4). Subsequent analysis showed that this signal had the same chromatographic behavior than CGA. These results suggest CGA is degraded which might be due to instability under the conditions used or by hydrolyzation through enzymes of A. alternata or by both processes. When after this degradation a certain low concentration is reached, the fungus starts to produce AOH. Under the conditions used here, this situation is reached after day 6. However depending on the actual conditions the inhibition time is variable. These results suggest that the degradation of CGA and not the adaptation of A. alternata to CGA, is the reason for the only transient ability of CGA to suppress AOH biosynthesis.
Influence of CGA on the expression of the pksJ gene
In order to analyse the influence of CGA on the expression of the pksJ gene, A. alternata BFE1316 was incubated on tomato agar plates as a model medium for the natural habitat with or without CGA for several days. Each day a sample was withdrawn and mRNA was isolated and used for a Real Time PCR expression analysis of the pksJ gene. The results are shown in Fig. 5. There is a clear difference between the AOH production kinetics by A. alternata on plain tomato medium or on medium with CGA. On plain tomato medium an early high expression of the pksJ gene became obvious. Already at days 4 and 5 a high optimum of expression was achieved which coincides with an early biosynthesis of AOH on a medium without added CGA (see above). In contrast, when the medium contains CGA also a quite high expression level is achieved, however only at day 6, which again is in agreement with the AOH production and the CGA degradation kinetics under these conditions (see above).
Taken together these results show an inhibitory influence of CGA on AOH biosynthesis at the transcriptional level. However apparently after degradation of CGA this inhibitory influence is relieved, transcription can start and AOH is produced.
Influence of CGA on the colonization of tomatoes
To analyse whether CGA indeed plays an inhibitory role during the infection of tomatoes by A. alternata, commercial tomatoes were wounded and were inoculated with A. alternata BFE1316 spores (103 spores/ml). As control, the plain spore solution was used. In optimization experiments it could be demonstrated that at a spore concentration of 103/ml, this strain of A. alternata showed a moderate colonization behaviour which enables the detection of differences. In contrast to this, higher spore concentrations lead to a complete colonization, disguising any difference. To analyse the influence of the polyphenols on colonization, the sample spore solutions contained 65, 650 μg/ml CGA, respectively. Each inoculation was performed several times. After inoculation the tomatoes were stored at 20 °C and a constant humidity for up to 6 days and the colonization was visually inspected each day. There was a clear and repeatable difference in the growth behaviour of A. alternata depending on the spore suspensions. The control showed a moderate colonization behaviour leading to a colonization of about two thirds of all wounds (Table 2). In contrast to this, CGA showed a concentration dependent inhibition of the colonization capacity of A. alternata. At 65 μg/ml, the CGA colonization was comparable to the control spore solution however at a concentration of 650 μg/ml the number of infected wounds were reduced to about one half of that of the control. Even the colony size of A. alternata was restricted compared to the control, indicating an inhibiting influence of CGA on colonization. Because it was formerly shown that AOH is a colonization factor for A. alternata, AOH was added to the CGA 650 μg/ml spore solution. Interestingly under these conditions an increase in the colonization capacity was observed, indicating that AOH partly overrides the inhibiting effect of CGA.
Discussion
It is a well-known fact that plants produce preformed substances, mainly secondary metabolites like essential oils, volatiles, saponines, jasmonates, glucosinolates and polyphenols (Osbourn 1996; Schena et al. 2008; Ahmad et al. 2013) but also proteins like glucosyl transferases, chitinases, β-glucanases or polygalacturonase inhibiting proteins (Santino et al., 2005) which protect them against fungal attack. One of the most important polyphenols in this respect is CGA. CGA occurs ubiquitously in plants and plant type products in high amounts, especially in the Solanaceae. It is one of the major phenolic compounds in certain fruits like peaches (Cheng and Crisosto, 1995) and prunes (Stacewicz-Sapuntzakis et al. 2001) and also tomatoes (Ruelas et al. 2006, Ahmad et al. 2013).
CGA is part of the plant defence system against fungi. Ruelas et al. (2006) analysed, whether fungal attack by A. alternata leads to changes in the concentration of CGA in tomato fruits. However, they only found a change in the concentration of vanillic acid but not CGA in tomatoes after infection with A. alternata. In contrast to these results, López-Gresa et al. (2011) found a rapid increase in the concentration of CGA in tomato plants after infection with the bacterial pathogen Pseudomonas syringae. Suárez-Quiroz et al. (2013) demonstrated a strong antifungal activity of CGA and various derivatives thereof on growth of several Aspergillus species, like A. flavus, A. nomius and A. parasiticus, as well as ochratoxigenic species like A. ochraceus and A. westerdijkiae. By means of an untargeted GC×GC-MS approach, CGA was identified as one of the major constituents of the secondary metabolome of ST, the resistant tomato genotype. Beside this difference, other variations in the metabolite profiles could also be identified between the two tomato genotypes. These variations in metabolites may also contribute to the differences in resistance of both tomato genotypes.
During the colonization of the tomato, the fungus has to overcome these defence mechanisms. This, for example, can be achieved by degradation of the respective inhibiting substance. The degradation of tomatine by fungal tomatinases is such an example (Osbourn 1996). On the other hand, the fungus can produce pathogenicity factors which weaken the response of the plant and support the infection by the fungus. Recently, various mycotoxins, secondary metabolites of the fungus, were shown to act as pathogenicity factors. This was shown for the trichothecenes produced by Fusarium (Desjardins et al. 1992), the patulin produced by P. expansum (Sanzani et al. 2012) or with alternariol biosynthesis in the case of A. alternata (Graf 2012). In all cases the fungus was no longer able to colonize the host plant, when the biosynthesis of the respective mycotoxin was inactivated by mutation. In case of A. alternata, an external addition of alternariol restored the pathogenicity (Graf et al. 2012). This situations suggests that each condition which reduces alternariol biosynthesis would reduce the pathogenicity and thereby the colonization capacity of A. alternata.
With the help of experiments on laboratory media, it could be demonstrated that increasing amounts of CGA reduced AOH biosynthesis in a concentration dependent manner. The biosynthesis of AOH was strongly inhibited at higher concentrations of CGA. The fact that only the production of AOH or its derivative AME, and not that of TeA was reduced indicated that CGA has a specific effect on AOH/AME biosynthesis. However, the inhibition of AOH biosynthesis is only temporal, due to the degradation of CGA. Under the conditions used here, the production of AOH starts after 6 days in the CGA containing culture compared to day 4 in the culture without CGA. This was also paralleled by a much earlier expression of the pksJ gene in the culture without CGA on tomato medium than in the CGA containing culture.
A chromatographic analysis revealed that the concentration of CGA reduces over time with a concomitant increase of AOH biosynthesis. These results suggest a degradation of CGA which might be due to the instability of the molecule or by degradation of CGA through A. alternata. Narita and Inouye (2013) demonstrated that CGA is derivatized in aqueous solutions in a pH dependent manner, but it was hypothesized that the derivates have equivalent biological activities. On the other hand Benoit et al. (2007) described a chlorogenic acid hydrolase in A. niger which is able to degrade CGA. It is possible that a similar enzyme is present in A. alternata. So apparently the time span of inhibition of AOH biosynthesis in A. alternata is dynamic and depends on the initial concentration of CGA and on the intensity of the CGA degrading activities. This dynamic balance between the presence of CGA and its degradation seems is apparently the reason the fact that the variability of the determined AOH and AME concentrations at day seven were higher in the CGA cultures compared to the control (Fig. 3a and b).
Under in vitro conditions the addition of CGA to the spore suspension of A. alternata inhibited colonization in a concentration dependent manner. According to Ruelas et al. (2006), CGA inhibits the germination of A. alternata spores which is one explanation for the defensive effect of CGA. Interestingly Graf (2012) could demonstrate that AOH has a positive effect on the germination of spores.
Taken together the outcome of an infection process apparently is dependent on the initial concentration of CGA in the tomato, which reduces AOH biosynthesis, and the degradation of CGA by a possible CGA hydrolase of A. alternate or by chemical means. If the CGA concentration is high enough, the colonization will be prevented, if, however, the degrading activity is high enough, AOH biosynthesis is relieved and colonization can be established.
References
Ackermann, Y., Curtui, V., Dietrich, R., Gross, M., Latif, H., Märtlbauer, E., & Usleber, E. (2012). Widespread occurrence of low levels of alternariol in apple and tomato products, as determined by comparative immunochemivcal assessment using monoclonal and polyclonal antibodies. Journal of Agricultural and Food Chemistry, 59, 6360–6368.
Ahmad, A., Shafique, S., & Shafique, S. (2013). Cytological and physiological basis for tomato varietal resistance against Alternaria alternata. Journal of the Science of Food and Agriculture, 93, 2315–2322.
Akamatsu, H., Ito, Y., Kodama, M., Otani, H., & Kohmoto, K. (1997). AAL-toxin-deficient mutants of Alternaria alternata tomato pathotype by restriction enzyme-mediated integration. Phytopathological, 87, 967–972.
Arie, T., Takahashi, H., Kodama, M., & Teraoka, T. (2007). Tomato as a model for plant-pathogen interactions. Plant Biotechnology, 24, 135–147.
Atanasova-Penichon, V., Pons, S., Pinson-Gadais, L., Picot, A., Marchegay, G., Bonnin-Verdal, M. N., Ducos, C., Barreau, C., Roucolle, J., Sehabiagoue, P., Carolo, P., & Richard-Forget, F. (2012). Chlorogenic acid and maize ear rot resistance: a dynamic study investigating Fusarium graminearum development, deoxynivalenol production, and phenolic acid accumulation. Molecular Plant—Microbe Interactions, 25, 1605–1616.
Benoit, I., Asther, M., Bourne, Y., Navarro, D., Canaan, S., Lesage-Meesen, L., Herweijer, M., Coutinho, P. M., Asther, M., & Record, E. (2007). Gene overexpression and biochemical characterization of the biotechnologically relevant chlorogenic acid hydrolase from Aspergillus niger. Applied and Environmental Microbiology, 73, 5624–5632.
Bostock, R. M., Wilcox, S., Wang, G., & Adaskaveg, J. E. (1999). Suppression of Monilia fructicola cutinase production by peach fruit surface phenolic acids. Physiology Molecular Plant Pathology, 54, 37–50.
Cheng, G. W., & Crisosto, C. H. (1995). Browning potential, phenolic composition, and polyphenoloxidase activity of buffer extracts of peach and nectarine skin tissue. Journal American Society Horticultural Science, 120, 835–838.
Desjardins, A. E., Hohn, T. M., & McCormick, S. P. (1992). Effect of gene disruption of trichodiene synthase on the virulence of Gibberella pulicaris. Molecular Plant—Microbe Interactions, 5, 214–222.
Graf E (2012) Vorkommen, Biodiversität und molekulares Monitoring von mykotoxinbildenden Alternaria Spezies in Lebensmitteln. Dissertion, Karlsruhe Institute of Technology (KIT)
Graf, E., Schmidt-Heydt, M., & Geisen, R. (2012) HOG MAP kinase regulation of alternariol biosynthesis in Alternaria alternata is important for substrate colonization. International Journal of Food Microbiology, 157, 353–359.
Horneburg, B., & Becker, H. (2011). Selection for Phytophthora field resistance in the F2 generation of organic outdoor tomatoes. Euphytica, 180, 357–367.
Logrieco, A., Moretti, A., & Solfrizzo, M. (2009). Alternaria toxins and plant diseases: an overview of origin, occurrence and risks. World Mycotoxin Journal, 2, 129–140.
López-Gresa, M. P., Torres, C., Campos, L., Lisón, P., Rodrigo, I., Bellés, J. M., & Conejero, V. (2011). Identification of defence metabolites in tomato plants infected by the bacterial pathogen Pseudomonas syringae. Environment Experimental Botany, 74, 216–228.
Moco, S., Bino, R. J., Vorst, O., Verhoeven, H. A., de Groot, J., van Beek, T. A., Vervoort, J., & de Vos, C. H. R. (2006). A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiology, 141, 1205–1218.
Mondello, L., Tranchida, P. Q., Dugo, P., & Dugo, G. (2008). Comprehensive two-dimensional gas chromatography–mass spectrometry: a review. Mass Spectrometry Reviews, 27, 101–124.
Morris, P. F., Connolly, M. S., & St. Clair, D. A. (2000). Genetic diversity of Alternaria alternata isolated from tomato in California assessed using RAPDs. Mycological Research, 104, 286–292.
Narita, Y., & Inouye, K. (2013). Degradation kinetics of chlorogenic acid at various pH values and effects of ascorbic acid and epigallocatechin gallate on its stability under alkaline conditions. Journal of Agricultural and Food Chemistry, 61, 966–972.
Osbourn, A. (1996). Saponins and plant defence—a soap story. Trends in Plant Science, 1, 4–9.
Osorio, S., Do, P. T., & Fernie, A. R. (2012). Profiling primary metabolites of tomato fruit with gas chromatography/mass spectrometry. Plant metabolomics Methods and Protocols, 860, 101–109.
Ostry, V. (2008). Alternaria mycotoxins: an overview of chemical characterization, producers, toxicity, analysis and occurrence in foodstuffs. World Mycotoxin Journal, 1, 175–188.
Ruelas, C., Tiznado-Hernández, M. E., Sánchez-Estrada, A., Robles-Burgueno, M. R., & Troncoso-Rojas, R. (2006). Changes in phenolic acid content during Alternaria alternata infection in tomato fruit. Journal Phytopathological, 154, 236–244.
Saha, D., Fetzner, R., Burkhardt, B., Podlech, J., Metzler, M., Dang, H., Lawrence, C., & Fischer, R. (2012). Identification of a polyketide synthase required for alternariol (AOH) and alternariol-9-methyl ether (AME) formation in Alternaria alternata. PloS One, 7, e40564.
Santino, A., Poltronieri, P., & Mita, G. (2005). Advances on plant products with potential to control toxigenic fungi: a review. Food Addictive Contamination, 22, 389–395.
Sanzani, S. M., Reverberi, M., Punelli, M., Ippolito, A., & Fanelli, C. (2012). Study on the role of patulin on pathogenicity and virulence of Penicillium expansum. International Journal of Food Microbiology, 153, 323–331.
Schena L, Nigro F, Ippolito A (2008) Natural antimicrobials to improve storage und shelf life of fresh fruits, vegetables and cut flowers, in Microbial Biotechnology in Horticulture, vol. 2, ed by Ray RC and Ward OP, CRC Press, Boca Raton, pp. 259–302.
Stacewicz-Sapuntzakis, M., Bowen, P. E., Hussain, E. A., Damayanti-Wood, B., & Farnsworth, N. R. (2001). Chemical composition and potential health effects of prunes: a functional food? Critical Reviews in Food Science, 41, 251–86.
Suárez-Quiroz, M. L., Alonso Campos, A., Alfaro, V., Gonzáles-Rios, O., Villeneuve, P., & Figueroa-Espinoza, M. C. (2013). Anti-Aspergillus activity of green coffee 5-O-caffeoyl quinic acid and its alkyl esters. Microbial Pathogenesis, 61–62, 51–56.
Terry, L. A., Joyce, D. C., Adikaram, N. K. B., & Kambay, P. B. S. (2014). Preformed antifungal compounds in strawberry fruit and flower tissues. Postharvest Biology and Technology, 31, 201–210.
Acknowledgments
We would like to thank Nicole Mischke and Marianne Urbanek for skillful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Wojciechowska, E., Weinert, C.H., Egert, B. et al. Chlorogenic acid, a metabolite identified by untargeted metabolome analysis in resistant tomatoes, inhibits the colonization by Alternaria alternata by inhibiting alternariol biosynthesis. Eur J Plant Pathol 139, 735–747 (2014). https://doi.org/10.1007/s10658-014-0428-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-014-0428-3