Abstract
The Generation R Study is a population-based prospective cohort study from fetal life until young adulthood. The study is designed to identify early environmental and genetic causes of normal and abnormal growth, development and health from fetal life until young adulthood. In total, 9,778 mothers were enrolled in the study. Prenatal and postnatal data collection is conducted by physical examinations, questionnaires, interviews, ultrasound examinations and biological samples. Major efforts have been conducted for collecting biological specimens including DNA, blood for phenotypes and urine samples. In this paper, the collection, processing and storage of these biological specimens are described. Together with detailed phenotype measurements, these biological specimens form a unique resource for epidemiological studies focused on environmental exposures, genetic determinants and their interactions in relation to growth, health and development from fetal life onwards.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Generation R Study is a population-based prospective cohort study from fetal life until young adulthood. The background and specific research projects of the study have been described in detail previously [1, 2]. Briefly, the Generation R Study is designed to identify early environmental and genetic causes of normal and abnormal growth, development and health from fetal life until young adulthood. The study focuses on four primary areas of research: (1) growth and physical development; (2) behavioural and cognitive development; (3) diseases in childhood; and (4) health and healthcare for pregnant women and children. The main outcomes and determinants are presented in Tables 1 and 2. The general aims of the study are:
-
(1)
To describe normal and abnormal growth, development and health from fetal life until young adulthood;
-
(2)
To identify biological, environmental and social determinants of normal and abnormal growth, development and health from fetal life until young adulthood;
-
(3)
To examine the effectiveness of current strategies for prevention and early identification of groups at risk.
For these aims, a detailed and extensive data collection has been conducted in the prenatal phase of the study and is currently conducted in the early postnatal period [2]. This data collection comprises also biological specimens including DNA, blood for phenotypes and urine samples. These biological specimens form the Generation R Study Biobank, which enables future epidemiological studies focused on environmental exposures, genetic determinants and their interactions in relation to growth, health and development from fetal life onwards. In this paper, the design, collection, processing and storage of these biological specimens in the prenatal phase of the study are described.
Design and biological samples collection
The Generation R Study is a population-based prospective cohort study from fetal life until young adulthood [2]. Mothers with a delivery date between April 2002 and January 2006 were eligible. Enrolment was aimed in early pregnancy but was allowed until birth of the child. Extensive assessments were carried out in mothers and their partners in pregnancy and are currently performed in their children. Assessments in pregnancy were planned in early pregnancy (gestational age <18 weeks), mid-pregnancy (gestational age 18–25 weeks) and late pregnancy (gestational age ≥25 weeks) and at birth. The individual time scheme of these assessments depended on the specific gestational age at enrolment [2]. The partners were assessed once in pregnancy. All children form a prenatally recruited birth-cohort that is currently being followed until young adulthood.
Additionally, more detailed assessments of fetal and postnatal growth and development are conducted in a randomly selected subgroup of Dutch children and their parents, referred to as the Generation R Focus Cohort. Studies conducted in this subgroup examine etiological associations using more time-consuming methods that cannot be applied in the whole cohort.
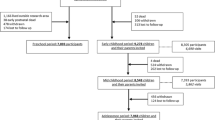
Biological materials have been collected in early, mid- and late pregnancy and at birth. Figure 1 shows the design of the collection of biological materials in this phase. In early, mid- and late pregnancy, biological materials were collected at the visits to one of our dedicated research centres. The planned amounts of blood taken by ante-cubital venous puncture were 35 ml in early pregnancy and 20 ml in mid-pregnancy from the mother and 10 ml from her partner. When mothers were enrolled in mid- and late pregnancy, 35 ml was taken at the first visit. Urine samples of mothers have been collected in early, mid- and late pregnancy from February 2004 until November 2005. Directly after delivery, midwives or obstetricians collected 30 ml cord blood from the umbilical vein.
The Generation R Study is approved by the Medical Ethical Committee of the Erasmus Medical Center, Rotterdam. Mothers and their partners received written and oral information about the study. Participants were asked for their written informed.
Study cohort
In total, 9,778 mothers were enrolled in the study. Of these mothers, 91% (n = 8,880) was enrolled in pregnancy. Only partners from mothers enrolled during pregnancy were invited to participate. In total, 71% (n = 6,347) of all partners were enrolled. The general characteristics of the mothers and their partners are presented in Table 3. Of all participating mothers, enrolment was in early pregnancy in 69% (n = 6,748), in mid-pregnancy in 19% (n = 1,857), in late pregnancy in 3% (n = 275) and at birth of their child in 9% (n = 898). The cohort comprises various ethnic groups. The largest ethnic groups are the Dutch, Surinamese, Turkish, Moroccan, Dutch-Antilles and Cape Verdian mothers.
Logistics
All biological samples were bar coded with a unique laboratory number. Blood or urine samples collected at one visit have the same bar code with an additional specific tube number. This combination forms a unique registration number. All following steps in processing, storing and data management of the samples are linked to this unique number.
All blood samples from the mother and partner were taken by research nurses and temporally stored at our research centre or one of the obstetric departments at room temperature for a maximum of 3 h. At least every 3 h, these blood samples were transported to a dedicated laboratory facility of the regional laboratory in Rotterdam, the Netherlands (STAR-MDC) for further processing and storage. Participants delivered their child either at home or at one of the hospitals in Rotterdam. The midwife or obstetrician collected cord blood samples. Subsequently, courier services with a 7-days and 24-h availability were responsible for transportation of these cord blood samples to our laboratory within 2 h.
After collection and transportation, all blood and urine samples have been centrally processed and stored at the STAR-MDC laboratory. Samples for DNA extraction have initially been stored as EDTA whole blood samples and subsequently transported to the Erasmus Medical Center for further processing.
All blood and urine samples are registered in Labosys (Philips) [3]. Both the anonymous person unique study numbers and all sample numbers are registered which enables matching of each sample to a study subject. Also, picking and defrosting of samples in registered in Labosys. A back-up of this registration is available at the Erasmus Medical Center.
Response rates
Response rates for collection of biological specimens are presented in Table 4. Overall response rates for blood collection in mothers are high. To get these high response rates, we have used the venous punctures that were already planned in routine care for additional blood sample collection when this was possible. Blood for DNA extraction is available in 91, 83 and 67% of the eligible mothers, partners and children. The larger number of missings in partners and children were mainly due to lack of consent and logistical constraints during delivery, respectively. As Table 4 shows, response rates for the various blood samples at one occasion were similar. Absolute numbers of urine samples are lower than the absolute number of blood samples because the urine sample collection was performed during a limited period in the prenatal phase of the study. Tables 5 and 6 show characteristics of children and partners with and without available DNA. In children, the number of missing blood samples in children with perinatal complications including low birth weight and preterm birth is higher because of the limited possibilities and priorities during these deliveries. At this moment, efforts are carried out for completing DNA collection in the children by buccal samples.
DNA
For DNA extraction, one 10 ml EDTA tube from mothers, partners and children at birth have been collected and stored directly as whole blood sample at −80°C. After completion of the blood sample collection, DNA extraction was started from white blood cells. First, DNA extraction from all children has been conducted manually using the Qiagen FlexiGene Kit (Qiagen Hilden, Germany). Currently, DNA extraction from 5 ml whole blood samples from the mothers and partners is performed by a Hamilton STAR multi-channel robot using AGOWA magnetic bead technology (at 72 samples per run) and is expected to be finished in spring 2008.
Extracted DNA is automatically collected in stock tubes (2D Matrix, Micronic) that are organized in 96-wells format, but that can be individually addressed. These stock samples are split into two tubes that are stored at different locations. One of the stocks will then be used for normalising the DNA concentrations in 96 deep well (DW) plates. This protocol, as well as other manipulations of the DNA samples are performed on a Caliper ALH3000 pipetting robot (8/96/384 channels) with a Twister module and a 96/384 wells Tecan GENios plus UV reader. In total, 90 DNA samples are normalised per 96 DW plate with 6 water blancs per plate. Additionally, a random selection of 5% of the total number of samples are put into separate 96 DW plates, again with 6 blancs per plate, for control purposes. For normalisation, sample and diluent volumes are automatically calculated and produced to obtain equal DNA concentrations for all samples (25–50 ng/μl). From these stock 96-DW plates, 384 DW plates are created, which comprise 24 blancs each. Subsequently, replica PCR plates (384 wells) with a concentration of 1 ng/μl are created for genotype studies.
Stock 96-well plates (DW and 2D Matrix, Micronic) are stored at −20°C at two different locations within the Erasmus Medical Center (Genetic Epidemiology Laboratory, Genetic Laboratory Internal Medicine). The 384 DW stock and PCR plates are stored at the Genetic Laboratory Internal Medicine, Erasmus Medical Center. All genotyping studies with Generation R DNA material are performed in-house at the high throughput genotyping facility in this Laboratory.
Genome Wide Association database
A new approach to identify most important common genetic variants that contribute to explain variance in phenotypes by a systematic analysis of our genome has only recently become available as the Genome Wide Association (GWA) analysis [4]. The GWA analysis is now generally considered a critical tool to identify genes for complex diseases and traits. The GWA analysis is based on genotyping large numbers (>500,000–2,000,000) of Single Nucleotide Polymorphisms (SNPs), and subsequently performing association analyses of the SNP genotypes (or haplotypes thereof) with the phenotype of interest. The power of GWA analyses has been recently demonstrated with the identification of susceptibility genes involved in a wide range of disorders varying from common and rare diseases including coronary heart disease, diabetes and multiple sclerosis [5–7]. These developments have led to a stream of novel disease genes, highlighting new etiological pathways and are expected to improve the understanding of the molecular basis of these diseases. Currently, we are preparing a GWA analysis in all children participating in the Generation R Study of whom DNA is available. For this, the infrastructure, used for the GWA analysis in Rotterdam Study, a population-based cohort study in about 15,000 adults in Rotterdam, the Netherlands is used [8].
Blood for phenotypes
Results of analyses performed in blood samples (5 ml EDTA plasma) for routine care for pregnant women (Hb, Ht, HIV, HBsAg, Lues, Rhesus factor and irregular antibodies) are obtained from midwife and obstetric registries.
All other EDTA plasma and serum samples from mothers and children have been processed and stored for future studies. Processing was aimed to be finished within a maximum of 3 h after venous puncture. Total processing time took 15 min. The samples were spun and the plasma and serum volume was distributed into 250 μl aliquots and transferred to 0.65 ml polypropylene tubes (Micronic) by a Tecan automatic liquid handler. From the mother, a set of on average 16 aliquots tubes (with a total volume of 4 ml) each from one plasma and serum sample were divided over four different microtiter trays. From the plasma and serum samples from the children, on average 11 tubes were filled with 250 μl aliquots (in total 2.75 ml) divided over three microtiter trays. The trays were immediately stored at −80°C. Each Micronic tube is uniquely coded on the bottom with the Traxis® 2D code.
The aliquots were divided over 4 trays of 96 tubes for storage in the freezers. Figure 2 shows the distribution of the samples in the 96 trays. All samples are stored at −80°C at one location (STAR-MDC laboratory) where they are distributed over four different freezers with different power supplies.
Urine
Urine samples (65 ml) were added to the data collection between February 2004 and November 2005. The urine samples were kept at room temperature for a maximum of 3 h and transported to the STAR-MDC laboratory for further processing. At the laboratory, the urine was filled out manually in one 5 ml and three 20 ml tubes. Before the urine was filled out, a gravity measurement was performed. The 5 ml urine tubes were send to the Department of Medical Microbiology of the Erasmus Medical Centre for Chlamydia analysis [9]. The remaining urine tubes (20 ml) were each stored at −20°C at the STAR-MDC laboratory.
Other population-based birth cohort studies
The Generation R Study is not the first birth-cohort study with an extensive collection of biological specimens. Compared to other birth-cohort studies, the size of the Generation R Study cohort is not larger but the measurements are more detailed and the study group has a large ethnic variety [10–14]. Studies having a similar design and biological specimens collection include the Avon Longitudinal Study of Parents and Children (ALSPAC) in Bristol, UK, the Danish National Birth Cohort Study, the Norwegian mother and child Cohort Study, the Southampton Women’s Survey (SWS) in Southampton, UK and The National Children’s Study in the USA.
Collaboration with these studies for funding, standardization, replication and quality management is necessary. Currently, we are exploring possibilities for formal collaboration.
Requests for biological samples use
The study has an open policy with regard to collaboration with other research groups. Request for collaboration and use of biological specimens should primarily be pointed to Albert Hofman (a.hofman@erasmusmc.nl). These requests are discussed in the Generation R Study Management Team regarding their scientific merits, study aims, overlap with ongoing studies, logistic consequences and financial contributions.
General policy is that collaborating researchers and groups are responsible themselves for the finances and that all laboratory tests are conducted in the Eramus Medical Center, Rotterdam. After approval of the project by the Generation R Study Management Team and the Medical Ethical Committee of the Erasmus Medical Center, the collaborative research project is embedded in one of the four research areas supervised by the specific principal investigator.
References
Hofman A, Jaddoe VW, Mackenbach JP, Moll HA, Snijders RF, Steegers EA, Verhulst FC, Witteman JC, Buller HA. Growth, development and health from early fetal life until young adulthood: the Generation R Study. Paediatr Perinat Epidemiol 2004;18:61–72.
Jaddoe VW, Mackenbach JP, Moll HA, Steegers EA, Tiemeier H, Verhulst FC, Witteman JC, Hofman A. The Generation R Study: design and cohort profile. Eur J Epidemiol. 2006;21:475–84.
Website: http://www.medical.philips.com/nl/products/medicallabit
Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 2005;6:95–108.
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–78.
Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS; Wellcome Trust Case Control Consortium (WTCCC), McCarthy MI, Hattersley AT. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–41.
International Multiple Sclerosis Genetics Consortium, Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 2007;357:851–6.
Hofman A, Breteler MM, van Duijn CM, Krestin GP, Pols HA, Stricker BH, Tiemeier H, Uitterlinden AG, Vingerling JR, Witteman JC.The Rotterdam Study: objectives and design update. Eur J Epidemiol. 2007;22:819–29.
Rours GI, Verkooyen RP, Willemse HF, van der Zwaan EA, van Belkum A, de Groot R, Verbrugh HA, Ossewaarde JM. Use of pooled urine samples and automated DNA isolation to achieve improved sensitivity and cost-effectiveness of large-scale testing for Chlamydia trachomatis in pregnant women. J Clin Microbiol. 2005;43:4684–90.
Jones RW, Ring S, Tyfield L, Hamvas R, Simmons H, Pembrey M, Golding J, ALSPAC Study Team. A new human genetic resource: a DNA bank established as part of the Avon longitudinal study of pregnancy and childhood (ALSPAC). Eur J Hum Genet. 2000;8:653–60.
Olsen J, Melbye M, Olsen SF, Sorensen TI, Aaby P, Andersen AM, et al. The Danish National Birth Cohort—its background, structure and aim. Scand J Public Health 2001;29:300–7.
Ronningen KS, Paltiel L, Meltzer HM, Nordhagen R, Lie KK, Hovengen R, Haugen M, Nystad W, Magnus P, Hoppin JA. The biobank of the Norwegian mother and child Cohort Study: a resource for the next 100 years. Eur J Epidemiol. 2006;21:619–25.
Branum AM, Collman GW, Correa A, Keim SA, Kessel W, Kimmel CA, et al. The National Children’s Study of environmental effects on child health and development. Environ Health Perspect 2003;111:642–6.
Inskip HM, Godfrey KM, Robinson SM, Law CM, Barker DJ, Cooper C. Cohort profile: The Southampton Women’s Survey. Int J Epidemiol 2006;35:42–8.
Acknowledgements
The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam. We gratefully acknowledge the contribution of general practitioners, hospitals, midwives and pharmacies in Rotterdam. The authors thank Claudia Kruithof, Jeannette Vergeer-Drop and Letty Jacobs for their assistance in data management. The first phase of the Generation R Study is made possible by financial support from the Erasmus Medical Center, Rotterdam, the Erasmus University Rotterdam and the Netherlands Organization for Health Research and Development (ZonMw).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Jaddoe, V.W.V., Bakker, R., van Duijn, C.M. et al. The Generation R Study Biobank: a resource for epidemiological studies in children and their parents. Eur J Epidemiol 22, 917–923 (2007). https://doi.org/10.1007/s10654-007-9209-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10654-007-9209-z