Abstract
Background
The long non-coding RNA X-inactive specific transcript (XIST) plays a crucial role in transcriptional silencing of the X chromosome. Zinc finger E-box-binding homeobox 1 (ZEB1) is a transcription factor involved in epithelial-mesenchymal transition (EMT) regulation.
Aims
This study aimed to investigate the impact of XIST on esophageal squamous cell carcinoma (ESCC) progression and its underlying mechanism involving the miR-34a/ZEB1/E-cadherin/EMT pathway.
Methods
XIST and ZEB1 expression were analyzed using quantitative PCR and immunohistochemistry. XIST knockdown was achieved in KYSE150 ESCC cells using siRNA or shRNA lentivirus transfection. Proliferation, migration, and invasion abilities were assessed, and luciferase reporter assays were performed to confirm XIST-miR-34a-ZEB1 interactions. In vivo ESCC growth was evaluated using a xenograft mouse model.
Results
XIST and ZEB1 were upregulated in tumor tissues, correlating with metastasis and reduced survival. XIST knockdown inhibited proliferation, migration, and invasion of KYSE150 cells. It decreased ZEB1 expression, increased E-cadherin and miR-34a levels. Luciferase reporter assays confirmed miR-34a binding to XIST and ZEB1. XIST knockdown suppressed xenograft tumor growth.
Conclusion
XIST promotes ESCC progression via the miR-34a/ZEB1/E-cadherin/EMT pathway. Targeting the XIST/miR-34a/ZEB1 axis holds therapeutic potential and serves as a prognostic biomarker in ESCC.
Graphical Abstract
This study has discovered an XIST-associated pathway involved in the regulation of ZEB1 expression, thereby influencing the progression of squamous cell carcinoma (ESCC). The specific mechanism is as follows: XIST can regulates ZEB1 expression by sponging miR-34a, ultimately promoting the metastasis of tumor by down-regulating the expression of E-cadherin.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seeing from the global perspective, esophageal cancer (EC), which includes esophageal squamous cell carcinoma (ESCC for short) and esophageal adenocarcinoma (EAC for short), is recognized as one of the most commonly-seen malignancies and the sixth leading culprit for cancer-related deaths [1, 2]. ESCC, the most common histological type of EC, accounts for more than 95% of all EC patient cases [3, 4]. Due to lack of early diagnostics, most ESCC patients are identified at the advanced stages with local tumor cells infiltration or metastasis, greatly weakening the treatment efficiency [4, 5]. Despite of great progress in treatment technologies, the 5-year survival rate of ESCC is still less than 20% because of recurrence and metastasis [6,7,8]. Therefore, it is important to further explore the underlying mechanisms of ESCC progression for developing more effective treatment of ESCC.
Tumor metastasis is a multi-step process, while epithelial-to-mesenchymal transition (EMT) is a major process accompanied by the loss of epithelial markers, such as E-cadherin, which leads to increasing invasive and metastatic abilities [9]. ZEB1, one factor of the zinc finger E-box-binding homeobox (ZEB) family, promotes tumor migration and invasion by accelerating EMT and inhibiting E-cadherin expression [10, 11]. Signal transduction and activation of ZEB1 is critical to cancer transformation and EMT [12]. Highly expressed ZEB1 shows noticeable associations with the malignancy of various cancers [12,13,14]. It was reported that ZEB1 expression by immunohistochemistry was identified in the cytoplasm (64.9% of cases), in nuclei (11.3% of cases) and in tumor stroma (80.1% of cases) of ESCC. Weak cytoplasmic expression of ZEB1 in ESCC was associated with longer survival [15]. In addition, LINC00152, functioning as an oncogenic lncRNA in various cancer types, was found to positively regulate ZEB1 expression through interaction with EZH2 to enhance EMT and oxaliplatin (L-OHP) resistance in EC cells [16]. Recently, the circular RNA Circ-ZDHHC5 was found to regulate the miR-217/ZEB1 axis and accelerate ESCC progression [17]. These reports support the idea that ZEB1 is generally believed to foster migration, invasion, and metastasis of tumors [12].
Long non-coding RNAs (lncRNA), a class of non-coding RNAs with a length of greater than 200 nucleotides, have long been regarded as crucial regulators for many cancers by playing a tumor oncogenic or suppressive role [18, 19]. LncRNA X-inactive specific transcript (XIST) functions as the master regulator of transcriptional silencing of X chromosome and has been demonstrated to be correlated with multiple cancers [20,21,22]. XIST has also been shown to be overexpressed in esophageal cancer tissues, and multiple molecular mechanisms, including regulation of the miR-101/EZHZ axis [23], sponging miR494 and regulating CDK6 expression [24], and sponging miR-129-5p and upregulating CND1 expression [25], have been reported to be involved in the roles of XIST in promoting the progressing of esophageal cancer. However, although these reports can partially explain XIST-mediated EC metastasis, the specific mechanisms underlying the link between XIST and ZEB1 have not been dissected.
In the present study, we confirmed the high expression level of XIST in human EC tissues and cell lines, and explored the possible interaction between XIST and its sponging miRNA (miR-34a), as well as the interaction between miR-34a and ZEB1 in EC cell line KYSE150. Through multiple in vitro molecular and cellular assays, we identified a novel XIST/miR-34a/ZEB1 pathway in EC cells, and verified the potential of targeted XIST knockdown for EC therapy in a KYSE150 tumor xenograft mouse model. Our study suggests that the molecules in the XIST/miR-34a/ZEB1 axis have the potential to serve as promising biomarkers for EC diagnosis, EC prognostication, and attractive targets for clinical therapy of EC..
Materials and Methods
Human ESCC Tissue Specimens
Samples from 66 patients were used in this study. All patients were surgically treated at The Fourth Hospital of Hebei Medical University (Hebei, China) between January 1, 2014 and December 31, 2015. Patient selection was as previously described [26]. Inclusion criteria for patient samples were as follows: 1, pathological diagnosis on initial gastroscopic bite: esophageal squamous cell carcinoma; 2, no preoperative history of radiotherapy; 3, preoperative assessment capable of R0 resection. Exclusion criteria for patient samples were as follows: 1, postoperative paraffin pathology was not esophageal squamous cell carcinoma; 2, patients who died within one month after surgery. All specimens were obtained and pathologically confirmed to be ESCC. Surgical samples were obtained within 30 min post-operative and immediately stored in liquid nitrogen.
The patients (52 males and 14 females) ranged in age from 50 to 87 years. The patients’ general data (gender, age, occupation, and family history) and data on tumor (pathological type, TNM stage and surgical situation) were extracted from medical records. Follow-up was performed according to the standard follow-up system of the hospital every 6 months after patients were discharged from the hospital. The deadline for the follow-up was December 31, 2019. The survival period was measured from the date of admission to the date of death or to the date of the follow-up deadline. During the follow-up period, 35 patients died due to ESCC, with no other underlying causes implicated. The clinical characteristics and outcomes of all participants are summarized in Table 1.
Immunohistochemical (IHC) Assay
Paraffin sections (4 mm thick) were deparaffinized and rehydrated, followed by treatment with 0.02 M EDTA buffer (pH 9.0, Gene Tech, USA). Then, the sections were immersed in 3% H2O2 and blocked with 5% normal goat serum in proper sequence, followed by incubation with monoclonal anti-ZEB1 antibody (1:500; Sigma, USA) overnight at 4 °C. The antibody was diluted in PBS buffer containing 5% normal goat serum. The negative control for each slide was incubated with 5% normal goat serum without the anti-ZEB1 antibody. The sections were then incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (ZSGB-BIO, China) for 45 min at 37 °C and revealed with diaminobenzidine tetrahydrochloride. The stained slides were scored by three pathologists who were unaware of the clinical diagnosis. The indexes of ZEB1 labeling were implemented so that samples were scored according to the percentage and intensity scores of positively stained tumor cells.
Cell Culture and Transfection
The ESCC cell lines were obtained from The Chinese National Infrastructure of Cell Line Resources (NICR; Beijing, China). Cells were routinely cultured in Roswell Park Memorial Institute (RPMI)-1640 supplemented with 10% Fetal Bovine Serum (FBS; Gibco, USA), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, USA) in the 37 °C incubator with 5% CO2.
KYSE150 cells were planted in plates about 16 h before transfection. The plasmids expressing control scramble shRNA and sh-XIST were prepared by Genehem (Shanghai, China). The sequences are listed below: scramble control shRNA, 5ʹ-TGGATCCAAGGTCGGGCAGGAAGAG-3ʹ; sh-XIST, 5ʹ-TCTCTGTCATTGCTTCTGTAGTCACAGTC-3ʹ. Transfection of KYSE150 cells was conducted with lipofectamine 2000 (Invitrogen, CA, USA) following the manufacturer’s instructions. Lentivirus for knocking down of miR-34a was ordered from Genehem (Shanghai, China), and the sequence cloned into the vectors is as follows: 5ʹ-TGCTGTTAGC TAATACTCAC AACAACGTTT TGGCCACTGA CTGACGTTGT TGTGTATTAG CTAA-3ʹ.
Cell Counting Kit-8 (CCK-8) Assays
Cell proliferation was measured by the CCK-8 assay kit (Dojindo Molecular Technologies, Inc.; Japan). Briefly, ESCC cells with the indicated gene expression manipulation were seeded in 96-well flat-bottom microplates with a density of approximately 5 × 103 cells/well, and incubated for 12, 24, 36, and 48 h, respectively. Then, 20 µL CCK-8 reagent was added into each well. Cells were incubated at 37℃ subsequently for another 2 h. The cell viability in each well was determined by measuring the optical absorbance at 450 nm on a microplate reader (Promega™; Madison, WI, USA).
Transwell Assays
For transwell assays, 2 × 104 cells were plated as scheduled in the top chamber and covered with or without membrane. Cells were seeded in the preset serum-free medium. In the meanwhile, serum-supplemented medium was used as an effective chemoattractant and put in the lower chamber. The cells were incubated for 24 h, and cells that had not migrated or invaded through the hole were gently eliminated with a cotton swab. After being fixed, the cells located on the lower surface of the membrane were then stained with Giemsa crystal violet solution and counted with the help of an optical microscope (TS100-F, Nikon, Japan).
Real-Time Quantitative PCR (qPCR)
Total RNA was extracted from tissue samples according to the instructions in the manual of iRNAVana™ PARISTM (Ambion, TX, USA). The levels of miRNAs were analyzed using qPCR with TaqMan, miRNA reverse transcription assays, and the appropriate primers according to the manufacturer's instructions. Briefly, 10 ng of total RNA was used as the template for 15 µL reverse transcription reactions. Probes were specially designed for specific mature miRNAs. For each miRNA, reactions were performed in triplicate using the 7500 RT-PCR system (Applied Biosystems), and RNU66 (Applied Biosystems, cat. no. 4373382) was used as the normalization control.
The sequences of primers used for qPCR assays are as follows: XIST, Forward, 5ʹ-TGGCTCTTCTTTCACGCTTT-3ʹ, Reverse, 5ʹ-TGGTGTCGTGGAGTCG-3ʹ;
miR-34a, Forward, 5ʹ-ACACTCCAGCTGTGACTGGTTGACCAGA-3ʹ, Reverse, 5ʹ-TCAACTGGTGTCGTGGA-3ʹ, RT, 5ʹ-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCCTCTG-3ʹ;
U6, Forward, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, Reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ; ZEB1, Forward, 5ʹ-TTCGAGCCATCATTAAAATCAC-3ʹ, Reverse, 5ʹ-CTGGGTGGTTCAGACTCACA-3ʹ; E-cadherin, Forward, 5ʹ-TGCCCAGAAAATGAAAAAGG-3ʹ, Reverse, 5ʹ-GTGTATGTGGCAATGCGTTC-3ʹ; GAPDH, Forward, 5ʹ-CGCTGAGTACGTCGTGGAGTC-3ʹ, Reverse, 5ʹ-GCTGATGATCTTGAGGCTGTTGTC-3ʹ.
Western Blotting Analysis
Cells were lysed with ice-cold lysis buffer (Keygen Biotech, China) for 30 min on ice. Cell lysates were then collected after centrifugation at 12,000 rpm for 5 min at 4 °C. The lysate protein at an amount of 60 µg was loaded, and the total cellular protein was separated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then transblotted overnight at 4 °C onto polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with antibody (anti-ZEB1, 1:1,000 dilution, Cell Signaling Technology (CST), USA; anti-E-cadherin, 1:2,000 dilution, CST, USA) at 4 °C overnight, washed three times with tris-buffered saline supplemented with 0.1% tween (TBST), and incubated with the IRDye® 800CW Conjugated Goat anti-Rabbit IgG (Catalog Number 926-32211, LI-COR Biosciences, USA) secondary antibody (1:2000 dilution) for 1 h at room temperature. The membranes were washed three times with TBST, and the immunoreactive bands were detected using the kit and the infrared (IR) laser-based instrumentation from LI-COR Biosciences, USA.
Luciferase Reporter Assay
The miRNA targets of XIST and the binding sites of miR-34a and XIST, as well as miR-34a and 3' untranslated region (UTR) of ZEB1 mRNA were predicted using the online tools starBase v 2.0 (http://starbase.sysu.edu.cn/starbase2/index.php). The DNA sequences with the predicted binding site regions and the corresponding mutated regions, were cloned into a luciferase-expressing vector psiCHECK2 (Shanghai Genechem, China). The luciferase reporter vectors containing the full length XIST or 3' untranslated region (UTR) of ZEB1 were constructed. Esophageal cancer cells were co-transfected with the indicated vectors or the control luciferase reporter vector together with miR-34a-mimic. At 48 h after transfection, cells were harvested and subjected to luciferase activity analysis using the Dual-Glo luciferase assay system (Shanghai Genechem, China) in accordance with the manufacturer’s instructions.
Xenograft Animal Model
BALB/c nude mice (4–5 weeks old, male) were purchased from Beijing Vital River Laboratory Animal Technologies Co. Ltd (Beijing, China). All animals were kept in a pathogen-free environment in the Animal Center of the Fourth Hospital of Hebei Medical University. and fed with food and water ad lib. Twelve nude mice aged 6 weeks were divided into two groups for cancer cells inoculation: siXIST and control (6 mice in each group). Stable XIST-knockdown KYSE150 cells were generated after infection of shXIST-expression lentivirus. A total of 5 × 106 stable XIST-knockdown KYSE150 cells (shXIST) or control KYSE150 cells (control) in 0.2 mL of phosphate buffered saline (PBS) (Corning, Manassas, V.A., USA) were subcutaneously injected into the right shoulder flank of nude mice. The xenograft size in each group was measured at 5, 10, 15, 20, 25, and 30 days after inoculation using a microcaliper, and the tumor volume was calculated according to the following formula: volume = 1/2 length × width2. At 30 days after tumor inoculation, mice were euthanized with CO2 inhalation and tumor tissues were harvested for additional analyses.
Statistical Analysis
Comparisons of the results with OS of patients were carried out using Kaplan–Meier log-rank testing. The correlation between the expression levels of XIST and ZEB in tumor tissues was evaluated using the COX regression analysis. Correlations between XIST level and patient characteristics were assessed using the χ2 statistical test. Data were analyzed by using SPSS 21.0 (SPSS Inc., Chicago, IL, USA) and are displayed as mean ± standard deviation (SD). A P < 0.05 was considered to be statistically significant.
Results
XIST and ZEB1 Are Overexpressed in ESCC and Their High Expressions Predict Worse Prognosis of ESCC Patients
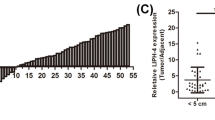
We evaluated the expression levels of XIST and ZEB1 in 66 paired ESCC and adjacent non-tumor tissues. We found that the XIST levels were significantly higher in tumor tissues (2.69 ± 1.71) than that in the paired adjacent non-tumor tissues (1.84 ± 1.67) (P < 0.001) (Fig. 1A). In addition, the ZEB1 protein expression levels as revealed by IHC staining were higher in tumor tissues (3.24 ± 2.79) than that in adjacent non-tumor tissues (1.06 ± 1.31) (P < 0.001) (Fig. 1B). We then analyzed XIST expression level in ESCC tissues and its effect on patient characteristics and survival time. Patients were categorized into two groups (high and low) based on the median level. The results showed that XIST expression was associated with postoperative metastasis (χ2 = 17.580, P < 0.001): a high level of XIST was correlated with postoperative metastasis (Table 2). Moreover, the ESCC patients with a high level of XIST had a significantly shorter overall survival (OS) and patients with a low XIST expression had a longer OS (Fig. 1C and Table 2). Additionally, patients were categorized into two groups (negative and positive) based on the IHC scores. We found that the expression of ZEB1 protein was associated with postoperative metastasis (χ2 = 6.110, P = 0.013) (Table 2). As expected, ESCC patients with ZEB1 positive expression had significantly shorter OS than the patients with negative ZEB1 expression (Fig. 1D and Table 2). Furthermore, the relative expression level of XIST in all examined tissues was positively correlated with the ZEB1 protein level by regression analysis (F = 58.529, P < 0.001) (Fig. 1E). Collectively, our results suggest that XIST and ZEB1 are overexpressed in ESCC and their high expressions predict worse prognosis of ESCC patients.
ESCC tumor tissues displayed high levels of XIST and ZEB1 expressions and their higher expression levels predicted worse prognosis. A The expression level of XIST in esophageal cancer tissues (T) and paired para-carcinoma tissues (N) was determined by qPCR. n = 66 for each group. B The expression level of ZEB1 protein in esophageal cancer tissues (T) and paired para-carcinoma tissues (N) was determined by IHC. Representative IHC images are shown on right. n = 66 for each group. C–D The relationship between XIST expression (C) or ZEB1 expression (D) in tumors and the prognosis of ESCC patients. The High and Low groups were determined based on the median values of XIST or ZEB1 expression. E Correlation between XIST and ZEB1 expression levels in ESCC cancer tissues
XIST Knockdown Inhibits the Proliferation, Migration, and Invasion of KYSE150 ESCC Cells, Which Can Be Attenuated by Further miR34a Silencing
To investigate the effects of XIST expression on ESCC disease progression, we sought to examine the impacts of manipulated expressions of XIST on cell proliferation and metastasis ability of human ESCC cells. We first evaluated the expression levels of XIST in several common human ESCC cell lines. Compared with the normal immortalized esophageal epithelial cell line NE1, the tested ESCC cell lines including ECA109, KYSE150, KYSE170, and TE1 displayed remarkably increased levels of XIST transcript. Particularly, KYSE150 cells had the highest expression level of XIST (Fig. 2A). Therefore, we selected the KYSE150 cell line as model cancer cells to further decipher the potential roles of XIST in the disease progression of ESCC. We confirmed successful knockdown of XIST, and the relative level of siXIST transfected cells is 0.38 in comparison to the control cells which had a relative level of 1 (Fig. 2B). Moreover, our proliferation assays demonstrated that KYSE150 cells with XIST knockdown had significantly decreased proliferation when compared with the control cells during the 3-day culturing, while a reduced proliferation by 39% was observed on Day 3 (Fig. 2C). miR-34a was reported as a sponge target of XIST [22]. Thus, we co-transfected miR-34-targeting siRNA into XIST knockdown cells, and found that these cells only showed a slightly decreased proliferation in comparison to the control cells on Day 3 (Fig. 2C). Furthermore, transwell assays showed that XIST knockdown significantly decreased the migration and invasion abilities of KYSE150 cells by 58% and 50%, respectively, while additional miR34a silencing remarkably restored the abilities of migration and invasion of XIST-knockdown KYSE150 cells (Fig. 2D). Therefore, XIST knockdown compromised the proliferation, migration, and invasion of KYSE150 ESCC cells, which could be attenuated by additional miR-34a silencing.
XIST knockdown-induced decrease proliferation, migration, and invasion of KYSE150 cells was attenuated by additional miR-34a silencing. A The expression levels of XIST in immortalized esophageal epithelial cells (NE1) and esophageal cancer cell lines were determined by qPCR. B Knockdown of XIST in KYSE150 cells was verified by qPCR. C–D KYSE150 cells were left untreated, or transfected with siXIST, or co-transfected with siXIST and simiR-34a, as indicated. The proliferation of indicated cells during a 3-day culturing period was measured by CCK-8 assays (C). The migration and invasion of the indicated KYSE150 cells were quantitated by transwell assays (D). Representative images are shown, and numbers of migrated/invaded cells per visual field were summarized. n = 3 for each group; *P < 0.05
XIST Regulates E-Cadherin and Indirectly Regulates ZEB1 Expression by Sponging miR-34a
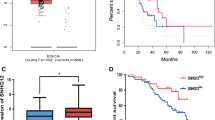
Knockdown of XIST reduced the migration and invasion of esophageal cancer cells, and E-cadherin is the key player in EMT that promotes tumor migration [9]. Therefore, we hypothesized that XIST may promote tumor migration by regulating E-cadherin expression. Quantitative assays showed that the level of E-cadherin mRNA was up-regulated by 62% (Fig. 3A) and expression of E-cadherin protein was increased by 360% upon knocking down of XIST (Fig. 3B). Additionally, we speculated that XIST might regulate E-cadherin expression through the miR-34a/ZEB1 axis based on previous study [27]. We thus verified the signal pathway and found that the level of miR-34a was significantly up-regulated while the level of ZEB1 mRNA was significantly decreased upon knocking down of XIST (Fig. 3A). A similar trend in changes of the protein levels for ZEB1 and E-cadherin was also identified in our western blot assays (Fig. 3B).
XIST knockdown increased E-cadherin expression and indirectly regulated ZEB1 expression through acting on miR-34a. A The levels of miR-34a, ZEB1 mRNA, and E-cadherin mRNA in control KYSE150 cells and siXIST-transfected KYSE150 cells were determined by qPCR. B The protein levels of ZEB1 and E-cadherin in control KYSE150 cells and XIST-knockdown KYSE150 cells were determined by western blot assays. Representative band images are shown, and relative protein levels were summarized. C–D The direct binding of XIST and miR-34a (C) or ZEB1 3’-UTR and miR-34a (D) was detected by Luciferase reporter assays. The luciferase activity in cells at 48 h after co-transfection of the indicated plasmids and miR-34a mimic was measured. n = 3 for each group; *P < 0.05
Next, luciferase reporter assays were performed to determine whether miR-34a specifically interacted with XIST and ZEB1 by binding to the predicted target sites.
Compared with the control group, miR-34a mimic significantly suppressed the luciferase activity of luciferase reporter vectors containing XIST (Fig. 3C) or 3’ UTR of ZEB1 (Fig. 3D), respectively. Whereas, the co-transfections of miR-34a mimic and mutated XIST or mutated 3’ UTR of ZEB1 resulted in largely unchanged luciferase activities, compared to the negative control (NC) co-transfections (Fig. 3C, D). These results indicate that miR-34a directly binds to the predicted target sites, and XIST can indirectly regulate ZEB1 expression by combining with miR-34a as a sponge.
XIST Knockdown Suppresses In Vivo Growth of the Xenografted ESCC Tumor
To test the potential for clinical translation of our in vitro findings, we set out to investigate whether silencing of XIST can facilitate in vivo tumor rejection in an ESCC xenograft animal model. We inoculated the control KYSE150 cells (Control) and the stable KYSE150 cells after infection of shXIST-expression lentivirus (siXIST group) into the right shoulder of nude mice, measured tumor growth, and euthanized the mice at 30 days after tumor inoculation. After 5 days, xenografted tumors grew to be palpable in all mice. As shown in Fig. 4A, the siXIST group at Day 30 had evidently smaller tumors in size. The volume of tumors in the siXIST group was significantly smaller than that in the Control group at the time of sacrifice (P = 0.012) (Fig. 4B). Protein quantitation assays showed that the relative expression level of the E-cadherin protein in the siXIST group was 3.12 folds of that in the Control group; while the relative expression of ZEB1 in the siXIST group was 6% of that in the control group (Fig. 4C). Taking together, these results are consistent with what we observed in the in vitro study: XIST silencing suppressed ESCC tumor growth.
Lentivirus-mediated knockdown of XIST inhibited in vivo tumor growth in nude mice. A–C Control KYSE150 cells (Control) and stable KYSE150 cells after lentivirus-mediated knockdown of XIST (siXIST) were subcutaneously injected into the right shoulder of nude mice. Xenografts were palpable 5 days after injection and all mice were sacrificed at 30 days after tumor cells inoculation. A The images of dissected tumors in each group. B Tumor volume was calculated at 5, 10, 15, 20, 25 and 30 days after tumors inoculation. The average xenograft volume of the siXIST group was significantly smaller than that of the control group. C The xenografts at Day 30 were dissociated for western blot analyses. The protein expressions of ZEB1 and E-cadherin in tumor tissues of two groups were determined. Representative band images are shown, and relative protein levels were summarized. n = 3 for each group; *P < 0.05
Discussion
ESCC remains one of the leading causes of cancer-related death [4, 5], and exploration of the complex molecular mechanisms underlying ESCC occurrence and development is still urgent. The oncogenic functions of the lncRNA XIST have been widely investigated in various cancers including ESCC [28]. Here, we identified a novel XIST/miR-34a/ZEB1 pathway in modulating the cell proliferation and metastasis of ESCC cells. We confirmed the higher expression levels of both XIST and ZEB1 protein in human ESCC tumor tissues and cell lines in comparison to the controls. Through multiple gene loss-of-expression assays, we validated the upstream and downstream interaction relationship between XIST and miR-34a, as well as between miR-34a and ZEB1 in KYSE150 cells. Notably, targeted down-regulation in XIST expressions in KYSE150 cells resulted in evidently slowed in vivo growth of xenografted ESCC tumors in nude mice, which was accompanied with increased E-cadherin expression and decreased ZEB1 expression in tumor tissues. These data unraveled a novel XIST-involved mechanism in ESCC development, and discovered new potential targets for effective therapy of ESCC.
The XIST gene is located in the Xq13.3 zone and is only expressed on the inactivated X chromosome [20]. The transcripts are long-chain noncoding RNA XIST and its antisense RNA TISX, which are not further translated into proteins [20]. XIST has been shown to be associated with multiple cancers and play different roles in different cancers. For example, XIST is down-regulated in breast cancer and plays a role in suppressing cancer. Loss of XIST expression can activate the MSN-c-Met pathway to promote breast cancer metastasis [21]. Our study and others suggest that XIST functions as an oncogenic gene in ESCC progression [23,24,25], as its downregulation suppresses the proliferation and metastasis potentials of ESCC cells. The similar functions of XIST were also observed in thyroid cancer: XIST is overexpressed in thyroid cancer tissues and cell lines, and promotes cell proliferation and tumor growth through the MET-PI3K-AKT signaling pathway [22]. Thus, the specific functions of XIST in cancer progression appear to be cell-type dependent, although in most cases XIST functions as an oncogene [28]. In addition, since XIST is involved in multiple aspects of carcinogenesis, including tumor onset, progression, and prognosis [28], the multilevel molecular functions of XIST in human tumors might be complicated. These cancer type-dependent roles of XIST in disease progression deserve more investigations to further reveal the molecular mechanisms underlying the divergences.
XIST can function as a sponge for a variety of endogenous miRNAs. For instance, XIST regulates the expression of E2F3 through competitively binding to miR-34a-5p, thereby playing the role of oncogene in nasopharyngeal carcinoma [29]. Additionally, XIST regulates RKIP expression by competitively binding to miR-23a, thereby exerting a tumor suppressive effect in prostate cancer [30]. This could help explain the distinct roles of XIST in promoting or inhibiting tumor progressing: the sponging miRNAs and the targets of miRNAs vary in different cellular settings. Moreover, XIST can also play a role in cancer progression by directly binding proteins. For example, XIST can directly bind to the EZH2 protein to epigenetically silence KLF2, thereby mediating its carcinogenic role in non-small cell lung cancer [31]. These studies proved that XIST could play an oncogenic role and may also play a role as tumor suppressor in different cancers, and these roles can be achieved through different interactions such as RNA-RNA and RNA–protein interactions.
In this study, we first determined the expression level of XIST in ESCC tissues and its relationship with clinical characteristics. We found that XIST was overexpressed in ESCC tissues and was associated with postoperative metastasis and prognosis. We further demonstrated the role of XIST in ESCC in vitro and found that knockdown of XIST in KYSE150 cells inhibited cell proliferation and metastasis potentials. Thus, we hypothesized that XIST may promote cancer progression by promoting the metastasis of esophageal cancer. As well known, E-cadherin is an EMT initiating factor and a tumor metastasis marker. Inhibition or loss of E-cadherin can activate tumor metastasis [32]. Our results show that XIST knockdown suppressed the metastasis potentials of esophageal cancer, which was associated with reduced E-cadherin expression and increased ZEB1 expression. ZEB1 is a key master factor controlling the structural network of transcription factors in EMT [12]. ZEB1 can recognize E-box sequences in the promoters of certain genes as a transcriptional repressor [12]. Consistent with our results, a negative correlation between ZEB1 and E-Cadherin expressions was identified in prostatic anomaly tissue [33] and in early zebrafish development [34]. These results suggest that XIST might promote the metastasis of ESCC through the ZEB1/E-cadherin pathway. Moreover, we found that ZEB1 was overexpressed in ESCC tissues and was associated with postoperative metastasis and prognosis. Since the expression level ZEB1 is positively correlated with the expression level of XIST in tumor tissues, it is plausible that XIST might enhance ESCC metastasis through ZEB1/E-cadherin-mediated EMT.
We further explored the mechanism of XIST in regulating ZEB1 and found that XIST, as a molecular sponge, competes with ZEB1 for binding to miR-34a, thereby releasing ZEB1 from binding with miR-34a. We reduced the expression level of miR-34a in the KYSE150 cells with XIST-knockdown and found that the decreases in cell proliferation, migration, and invasion ability were restored upon silencing of miR-34a. In addition, our luciferase reporter assays supported that miR-34a directly binds to the predicted target site of XIST and 3’UTR of ZEB1. Therefore, XIST indirectly regulates ZEB1 expression by combining with miR-34a as a sponge. Since miR-34a silencing helps restore the malignant behavior of ESCC, miR-34a functions as a tumor suppressor in our study. In line with this notion, miR-34a was reported to inhibit the progression of human esophageal cancer in multiple studies [35,36,37,38]. Although these reports proposed distinct mechanisms underlying the roles of miR-34a in inhibiting tumor metastasis, we are the first to reveal the role of miR-34a as a bridge to link the functions of XIST and ZEB1/ECM.
Conclusions
In summary, for the first time, we identified a novel XIST/miR-34a/ZEB1 pathway that is implicated in modulating cell proliferation and metastasis potentials of human esophageal squamous cell carcinoma both in vitro and in vivo. Ours and others’ studies suggest that the oncogenic lncRNA XIST is not only a biomarker for diagnosis and prognosis analyses of ESCC, but also a therapeutic target. Our work suggests that the approaches aimed at downregulating the expressions of XIST/ZEB1 and downregulating miR-34a expression in tumor are promising treatment strategies for ESCC therapy.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
References
Arnold M, Soerjomataram I, Ferlay J et al. Global incidence of oesophageal cancer by histological subtype in 2012. Gut 2015;64:381–387.
Ferlay J, Soerjomataram I, Dikshit R et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–E386.
Anvari K, Sima HR, Seilanian Toussi M et al. EGFR Expression in Patients with Esophageal Squamous Cell Carcinoma and its Association with Pathologic Response to Preoperative Chemoradiotherapy: A Study in Northeastern Iran. Arch Iran Med 2017;20:240–245.
Reichenbach ZW, Murray MG, Saxena R et al. Clinical and translational advances in esophageal squamous cell carcinoma. Adv Cancer Res 2019;144:95–135.
Hirano H, Kato K. Systemic treatment of advanced esophageal squamous cell carcinoma: chemotherapy, molecular-targeting therapy and immunotherapy. Jpn J Clin Oncol 2019;49:412–420.
Zeng H, Zheng R, Guo Y et al. Cancer survival in China, 2003–2005: a population-based study. Int J Cancer 2015;136:1921–1930.
Domper Arnal MJ, Ferrandez Arenas A, Lanas Arbeloa A. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol 2015;21:7933–7943.
Nagami Y, Ominami M, Shiba M et al. The five-year survival rate after endoscopic submucosal dissection for superficial esophageal squamous cell neoplasia. Dig Liver Dis 2017;49:427–433.
Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med 2018;12:361–373.
Qin Y, Tang B, Hu CJ et al. An hTERT/ZEB1 complex directly regulates E-cadherin to promote epithelial-to-mesenchymal transition (EMT) in colorectal cancer. Oncotarget 2016;7:351–361.
Li X, Gao D, Wang H et al. Negative feedback loop between p66Shc and ZEB1 regulates fibrotic EMT response in lung cancer cells. Cell Death Dis 2015;6:e1708.
Caramel J, Ligier M, Puisieux A. Pleiotropic Roles for ZEB1 in Cancer. Cancer Res 2018;78:30–35.
Shao Q, Wang Q, Wang J. LncRNA SCAMP1 regulates ZEB1/JUN and autophagy to promote pediatric renal cell carcinoma under oxidative stress via miR-429. Biomed Pharmacother 2019;120:109460.
Zhao L, Liu Y, Zhang J et al. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis 2019;10:731.
Goscinski MA, Xu R, Zhou F et al. Nuclear, cytoplasmic, and stromal expression of ZEB1 in squamous and small cell carcinoma of the esophagus. APMIS 2015;123:1040–1047.
Zhang S, Liao W, Wu Q et al. LINC00152 upregulates ZEB1 expression and enhances epithelial-mesenchymal transition and oxaliplatin resistance in esophageal cancer by interacting with EZH2. Cancer Cell Int 2020;20:569.
Wang Q, Yang L, Fan Y et al. Circ-ZDHHC5 Accelerates Esophageal Squamous Cell Carcinoma Progression in vitro via miR-217/ZEB1 Axis. Front Cell Dev Biol 2020;8:570305.
Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661–5667.
Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965–3981.
Brown CJ, Lafreniere RG, Powers VE et al. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 1991;349:82–84.
Xing F, Liu Y, Wu SY et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res 2018;78:4316–4330.
Liu H, Deng H, Zhao Y et al. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res 2018;37:279.
Wu X, Dinglin X, Wang X et al. Long noncoding RNA XIST promotes malignancies of esophageal squamous cell carcinoma via regulation of miR-101/EZH2. Oncotarget 2017;8:76015–76028.
Chen Z, Hu X, Wu Y et al. Long non-coding RNA XIST promotes the development of esophageal cancer by sponging miR-494 to regulate CDK6 expression. Biomed Pharmacother 2019;109:2228–2236.
Wang H, Li H, Yu Y et al. Long non-coding RNA XIST promotes the progression of esophageal squamous cell carcinoma through sponging miR-129-5p and upregulating CCND1 expression. Cell Cycle 2021;20:39–53.
Patino CM, Ferreira JC. Inclusion and exclusion criteria in research studies: definitions and why they matter. J Bras Pneumol 2018;44:84.
Xu Y, Guo B, Liu X et al. miR-34a inhibits melanoma growth by targeting ZEB1. Aging (Albany NY) 2021;13:15538–15547.
Yang J, Qi M, Fei X et al. Long non-coding RNA XIST: a novel oncogene in multiple cancers. Mol Med 2021;27:159.
Song P, Ye LF, Zhang C et al. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene 2016;592:8–14.
Du Y, Weng XD, Wang L et al. LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget 2017;8:94358–94370.
Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun 2016;478:811–817.
Wong SHM, Fang CM, Chuah LH et al. E-cadherin: Its dysregulation in carcinogenesis and clinical implications. Crit Rev Oncol Hematol 2018;121:11–22.
Pratiwi SE, Wahyuningrum SN, Putri RP et al. ZEB1 is Negatively Correlated with E-Cadherin in Prostatic Anomaly Tissue. Molecular and Cellular Biomedical Sciences 2022;6:28–34.
Vannier C, Mock K, Brabletz T et al. Zeb1 regulates E-cadherin and Epcam (epithelial cell adhesion molecule) expression to control cell behavior in early zebrafish development. J Biol Chem 2013;288:18643–18659.
Shi H, Zhou S, Liu J et al. miR-34a inhibits the in vitro cell proliferation and migration in human esophageal cancer. Pathol Res Pract 2016;212:444–449.
Cui XB, Peng H, Li RR et al. MicroRNA-34a functions as a tumor suppressor by directly targeting oncogenic PLCE1 in Kazakh esophageal squamous cell carcinoma. Oncotarget 2017;8:92454–92469.
Zuo J, Zhu K, Wang Y et al. MicroRNA-34a suppresses invasion and metastatic in esophageal squamous cell carcinoma by regulating CD44. Mol Cell Biochem 2018;443:139–149.
Zhou H, Yang L, Xu X et al. miR-34a inhibits esophageal squamous cell carcinoma progression via regulation of FOXM1. Oncol Lett 2019;17:706–712.
Acknowledgments
None.
Funding
None.
Author information
Authors and Affiliations
Contributions
BG and MM carried out the studies, participated in collecting data, and drafted the manuscript. JJ and GT performed the statistical analysis and participated in its design. MH and ZT participated in acquisition, analysis, or interpretation of data and draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Institutional Human Ethics Committee of Fourth Hospital of Hebei Medical University (human study ethics approval number 2021KY047), and prior informed consent was obtained from all the patients. The procedures for care and use of animals were approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University (animal study ethics approval number 2020KY135) and all applicable institutional and governmental regulations concerning the ethical use of animals were followed.
Consent for publication
Not applicable.
Competing interest
The authors declare no potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Guo, B., He, M., Ma, M. et al. Long Non-coding RNA X-Inactive Specific Transcript Promotes Esophageal Squamous Cell Carcinoma Progression via the MicroRNA 34a/Zinc Finger E-box–Binding Homeobox 1 Pathway. Dig Dis Sci 69, 1169–1181 (2024). https://doi.org/10.1007/s10620-024-08269-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-024-08269-0