Abstract
Background
MicroRNAs, targeting mRNAs of cancer-associated genes, are often aberrantly expressed in human gastric cancer (GC).
Aim
We have examined the possible role and mechanisms of miRNA regulation of Prdx-6 in the development and progression of H. pylori-related gastric mucosal lesions.
Methods
First, miR-24-3p was predicted to target Prdx-6, and this negative regulation was validated by luciferase reporter analyses, Western blot, and quantitative RT-PCR. Next, immunohistochemistry and in situ hybridization were performed to detect the Prdx-6 and miR-24-3p expression in tissue microarrays of gastric mucosal lesions. Finally, the miR-24-3p function in GC cell line N87 was examined by MTT, Annexin V-FITC, PI, transwell migration, and Matrigel invasion assays.
Results
In our study, Prdx-6 expression was negatively regulated by miR-24-3p expression and miR-24-3p interacted with the 3′-untranslated region of Prdx-6 to down-regulate its expression level. In addition, miR-24-3p expression gradually decreased in human gastric specimens from chronic superficial gastritis (CSG) to dysplasia and was upregulated in GC tissues compared with adjacent normal tissues. Contrary to this, Prdx-6 expression showed inverse tendency in the same tissue. More so, expression of miR-24-3p was reduced in samples with H. pylori infection, especially in CSG. Moreover, miR-24-3p was associated with GC lymph nodes and liver metastasis. Gain- or loss-of-function experiments showed that miR-24-3p significantly inhibited N87 cell growth, migration, and invasion and promoted apoptosis, while Prdx-6 reversed these miR-24-3p effects.
Conclusions
miR-24-3p was identified as a regulator of development and progression of H. pylori-related gastric mucosal lesions.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a novel class of endogenous, small noncoding RNAs that negatively regulate gene expression by perfect or imperfect base pairing with the 3′-untranslated region (UTR) of mRNAs [1]. MiRNAs play important roles in numerous biological processes, including cell development, apoptosis, and proliferation [2, 3]. Aberrant expression of miRNAs is involved in various cancers, including gastric cancer (GC), which is the second leading cause of tumor-related deaths worldwide [4]. Therefore, revealing the role of miRNAs in gastric cancer tumorigenesis may provide novel targets for prevention and treatment. Peroxiredoxins (Prdxs) are a family of small proteins that catalyze the reduction in peroxides using their conserved cysteine (Cys) residues as catalytical centers [5, 6]. Peroxiredoxin 6 (Prdx-6), belonging to the class of 1-Cys Prdx, plays an important role in neutrophil function and supports the optimal activity of Nox2 [7].
In our previous study, it was shown that Prdx-6 was associated with the development and progression of H. pylori-related gastric mucosal lesions [8]. In addition, bioinformatics prediction indicated that there are five miRNAs that might regulate Prdx-6 expression level (hsa-miR-24, hsa-miR-186, hsa-miR-149, hsa-miR-124, and hsa-miR-506). Based on the integrated analysis of copy number variations (CNVs), genomic alterations, miRNA expression profiling, microRNA 24 drew our attention and it has two gene subtypes—miR-24-3p, miR-24-5p [9]. After preliminary experiments, miR-24-3p was found to be highly expressed in GC. In this study, it is hypothesized that miR-24-3p might influence the development and progression of gastric mucosal lesions as well as tumorigenic properties of GC cell lines via Prdx-6.
Materials and Methods
Human Tissue Specimens and Tissue Microarray
Paraffin-embedded tissue samples, including 35 chronic superficial gastritis (CSG) samples, 43 chronic atrophic gastritis (CAG) samples, 41 intestinal metaplasia (IM) samples, 39 dysplasia (Dys) samples, 31 human GC samples, and 31 samples from adjacent normal mucosal tissues, were included in this study. Resected tissues from 31 patients with GC were harvested at the Department of Surgery, while other specimens were collected from patients undergoing gastroscopy at the Department of Gastroenterology, Aerospace Center Hospital, from 2005 through 2012. None of the GC patients had received adjuvant chemotherapy before surgery. All samples were verified by pathological examination and were used for a tissue microarray. Clinicopathological data were reviewed, and TNM staging classification was based on the American Joint Committee on Cancer criteria (UICC/AJCC, 7th edition). Written informed consent from the donor was obtained for the use of this sample in research. The study was approved by the Ethics Committee of Aerospace Center Hospital (20140627YJS-01).
Cell Lines and Cell Culture
All cell lines were preserved in our laboratory, and these were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY, USA.) with 10 % fetal bovine serum (FBS) (Invitrogen, Grand Island, NY, USA) and incubated in a humidified incubator (5 % CO2) at 37 °C.
Plasmid Construction and Cell Transfection
The expression plasmids containing miR-24-3p and Prdx-6 sequences were created by polymerase chain reaction (PCR) amplification using human genomic DNA as a template. The primers used were as follows: miR-24-3p-F, 5′-GATCCTGGCTCAGTTCAGCAGGAACAGC-3′, and miR-24-3p-R, 5′-TCGAGCTGTTCCTGCTGAACTGAGCCAG-3′; Prdx-6-F, 5′-TAGCGTTTAAACTTAAGCTTATGCCCGGAGGTCTGCTTCTCGGGG-3′, and Prdx-6-R, 5′-ACGGGCCCTCTAGACTCGAGTTAAGGCTGGGGTGTGTAGCGGAGG-3′. The PCR products were cloned into the pcDNA3.1(−) expression vector (Invitrogen, Grand Island, NY, USA) and confirmed by DNA sequencing. The empty vector was used as a control.
N87 cells were seeded at 1000 cells per well in six-well plates. Cells were transfected with miR-24-3p or empty vector using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA). Twenty-four hours after transfection, the transfected cells were incubated for 3 weeks in DMEM with 400 μg/ml of G418 (Glenview, IL, USA). Next, Prdx-6 expression vectors were transiently transfected into N87 cells overexpressing miR-24-3p or N87 cells transfected with an empty vector.
The hsa-miR-24-3p mimics, inhibitor, and their negative controls were purchased from Invitrogen (Shanghai, China). Oligonucleotide transfection was performed with Lipofectamine 2000 reagent at a final concentration of 100 nmol/l.
Luciferase Reporter Assay
To generate the 3′-UTR luciferase reporter, the full length of the Prdx-6 3′-UTR sequence was cloned into the downstream region of the firefly luciferase gene using the pmirGLO Dual-luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA; E1910). Mutant miR-24-3p target sites in the Prdx-6 3′-UTR sequence were used as corresponding controls. N87 and AGS Cells were seeded in triplicate in 24-well plates and cultured for 24 h. Luciferase reporter vectors were cotransfected with miR-24-3p mimic or negative control (NC) using Lipofectamine 2000. Luciferase activities were measured 24 h after transfection using the Dual-Luciferase Reporter Assay System (Promega). Two independent experiments were performed in triplicate. Firefly luciferase activity was normalized against Renilla luciferase activity.
Quantitative Reverse Transcription PCR (RT-qPCR)
Total RNA was extracted from cells using a standard Trizol protocol (Invitrogen). Reverse transcription (RT) reactions were carried out using EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China), and stem-loop RT primer for miR-24-3p was: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGTTC. The miRNA expression level was evaluated using the miRNA Q-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA) according to the manufacturer’s protocol. Expression of miR-24-3p and Prdx-6 was detected using the SYBR Green method, and primers for miR-24-3p and Prdx-6 were: 5′-TTTGGCTCAGTTCAGCAG-3′ (F), 5′-TTTGGCACTAGCACATT-3′ (R) and 5′-GGATGGGGATAGTGTGATGG-3′ (F), 5′-CTGACATCCTCTGGCTCACA-3′ (R).The results were normalized against U6 and β-actin.
Western Blot Analysis
Total proteins were extracted from cell lines in lysis buffer containing 50 mM Tris (pH 6.8), 100 mM dithiothreitol, 1 % sodium dodecyl sulfate, and 25 % glycerol. Equal amount of proteins were electrophoresed by 12 % SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride (PVDF) membranes using the Mini-PROTEAN 3 system (Bio-Rad, Hercules, USA). PVDF membranes were blocked in 5 % bovine serum albumin and incubated with rabbit antihuman Prdx-6 polyclonal antibody (1:2000; Abcam, Cambridge, MA, USA) followed by horseradish peroxidase-linked goat–antimouse IgG (Proteintech, Chicago, IL, USA). β-Actin was used as a loading control (Sigma, St. Louis, MO, USA). Protein intensity was quantified using the LANE 1D Analyzer software (Sage Creation, Beijing, China).
Immunohistochemistry (ICH) and In Situ Hybridization (ISH)
Tissue microarrays were stained with rabbit antihuman Prx-6 polyclonal antibody (1:1000; Abcam, Cambridge, MA, USA). IHC staining was performed using Envision kit (Dako, Glostrup, Denmark).
The sequence of the miR-24-3p probe was 5′-CTGTTCCTGCTGAACTGAGCCA-3′ (miRCURY ™ probe, Exiqon, Vedbaek, Denmark). A U6 probe (sequence: CACGAATTTGCG TGTCATCCTT; Exiqon) was used as the positive control. ISH was implemented according to the guidelines stated in the miRCURY LNA microRNA ISH Optimization Kits (FFPE) protocol (Exiqon). Slides were treated with proteinase K at 15 µg/ml for 10 min at 37 °C. Hybridization was performed at 62 and 58 °C, respectively, for the following: 5′-digoxigenin (DIG) labeled (U6) and double DIG (miR-24-3p), LNA-modified oligonucleotide ISH probes.
MTT Assay
N87 transfected and control cells were seeded in 96-well plates with 1 × 104 cells per well in 100 µl of cell culture medium, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltertrazolium bromide (MTT) was added at 0, 24, 48, 72, and 96 h. After 4 h incubation at 37 °C in 5 % CO2, 200 µl of dimethyl sulfoxide (DMSO) was added to solubilize the formazan product. The absorbance at 570 nm was determined using a microplate reader (Bio-Rad, Hercules, USA).
Apoptosis Analysis
The Annexin V-FITC Apoptosis Detection Kit (Beijing Biosea Biotechnology Co., Ltd., Beijing, China) was used to examine cell apoptosis. In brief, trypsinized cells were double-stained with fluorescein isothiocyanate (FITC)-conjugated Annexin V (20 μg/ml) and propidium iodide (PI; 50 μg/ml) according to the manufacturer’s protocol. Cells (1x104 per well) were detected by FACSorter flow cytometer (BD, New Jersey, USA), and data were analyzed using FASCDiva software. Cells in the early stages of apoptosis were Annexin V-positive and PI-negative, whereas cells in the late stages of apoptosis were both Annexin V- and PI-positive.
Migration and Invasion Assay
For the migration assay, cells at a concentration of 5 × 104 cells/ml were plated onto six-well transwell plates (Corning, Tewksbury, MA, USA) with an 8.0-μm pore polycarbonate membrane. For the invasion assay, cells (5 × 104 cells/ml) were plated onto six-well transwell plates pre-coated with Matrigel (BD, Shanghai, China). In both assays, DMEM containing 10 % FBS was used as a chemoattractant and was added to the lower chamber. After 24 and 48 h, membranes were fixed with 95 % alcohol for 20 min and stained with hematoxylin for 10 min. Six random fields for each insert were counted, and the results were averaged among three independent experiments.
Statistical Analysis
All of the enumerated data were analyzed by the χ 2 and Fisher’s exact tests, and comparisons of the continuous data between two groups were analyzed using the Student’s t test when there were only two groups. Data are expressed as the mean ± SD or percentage and constituent ratio. All statistical analyses were performed using SPSS software (version 17.0, IBM, USA) and P < 0.05 or P < 0.05/10 (Bonferroni correct P value) was considered statistically significant.
Results
miR-24-3p Was Identified as an Upstream miRNA of Prdx-6 by miRNA Selection
For the selection of miRNAs potentially targeting Prdx-6 to be included in subsequent analysis, TargetScan 6.2 software (http://genes.mit.edu/targetscan/index.html) was used. Based on the June 2012 release version, five microRNAs were found that potentially targeted Prdx-6. Next, an integrated analysis was performed, which included examination of publicly available data on miRNA expression profiling, CNVs, genomic alterations [8], and in our preliminary experiments, miR-24-3p was identified as an upstream miRNA of Prdx-6.
Negative Regulation Between miRNA-24-3p and Prdx-6 Expression Is Present in Gastric Cancer Cells
To investigate the expression of miRNA-24-3p and Prdx-6, RT-qPCR and Western blot were performed in five types of GC cell lines (BGC823, MGC803, SGC7901, AGS, and N87) and one immortalized normal gastric mucosal epithelial cell line (GES-1). Compared with GES-1, the level of miRNA-24-3p expression was elevated in the GC cell lines, especially in BGC823 and MGC803 (Fig. 1a). Western blot and RT-qPCR analysis revealed that Prdx-6 expression displayed the opposite tendency as miRNA-24-3p expression (Fig. 1b). To further examine this possible negative relation between miRNA-24-3p and Prdx-6, transfected SGC7901, N87, AGS, and GES-1 (cell lines with low miRNA-24-3p expression) were transfected with miRNA-24-3p mimics and NC mimics. Cell lines with higher miRNA-24-3p expression (BGC823 and MGC803 cells) were transfected with miRNA-24-3p inhibitor and NC inhibitor and, after 24 h, examined for expression of Prdx-6. Compared with the NC group, cells transfected with inhibitor had a 4.45-fold (BGC823) and 2.46-fold (MGC803) increase in Prdx-6 expression (Fig. 1c). In cells transfected with mimics, a decrease in Prdx-6 expression was present in addition to SGC7901 (Fig. 1d). The analysis of the Prdx-6 expression in different transfected cell lines showed a negative regulation between miRNA-24-3p and Prdx-6 expression.
Negative regulation between miR-24-3p and Prdx-6 expression. a miR-24-3p expression in five GC cell lines and GES-1. miR-24-3p mRNA levels were detected by RT-qPCR, compared with GES-1, and the level of miR-24-3p expression was elevated in GC cells, especially in BGC823 and MGC803 cell lines. b Expression of Prdx-6 in six cell lines. Prdx-6 protein and mRNA level were detected by RT-qPCR and Western blot analysis, respectively. Prdx-6 expression displayed the opposite tendency to miRNA-24-3p expression when examined in the same cell line. c Prdx-6 expression in BGC823 and MGC803 cells was analyzed 24 h after transfection with inhibitor and NC inhibitor. Inhibition of endogenous miR-24-3p caused an increase in Prdx-6 expression. d Prdx-6 expression was repressed by miR-24-3p in N87, AGS, and GES-1 cells. Cells were transfected with miRNA-24-3p mimics or NC mimics; 24 h later, the Prdx-6 expression levels were examined by RT-PCR and Western blot analysis. Relative Prdx-6 expression was normalized against β-actin, and data in histograms are mean relative quantity. Mean RQ and SD are shown in each figure, and Student’s t test with SPSS (version 11.0, IBM, USA) showed a *P < 0.05
Prdx-6 Is a Direct Target of miR-24-3p in Gastric Cancer Cells
Though Prdx-6 has been regarded as a target of miRNA-24-3p, it is unclear whether miRNA-24-3p can directly recognize the 3′-UTR of Prdx-6 mRNA. According to the predicted target site from TargetScan, two luciferase reporters were constructed: one containing a wild-type Prdx-6 3′-UTR sequence that contains the miR-24-3p binding site (pGL3-wt-3′-UTR) and another containing a mutated Prdx-6 3′-UTR (pGL3-mt-3′-UTR) (Fig. 2a). Next, luciferase reporter assays were performed to verify a direct interaction between miR-24-3p and the 3′-UTR of Prdx-6. The luciferase reporter constructs were transfected into N87 and AGS cells, along with miR-24-3p mimics or negative control oligos. Compared to the NC, the relative luciferase activity of co-transfection with miR-24-3p mimics and the pGL3-wt-3′-UTR vector decreased. However, miR-24-3p mimics did not have any effect on luciferase activity when the cells were transfected with pGL3-mt-3′-UTR vector (Fig. 2b). These results suggest that the Prdx-6 gene is one of the direct targets of miR-24-3p, and miR-24-3p negatively regulates Prdx-6.
Prdx-6 is a direct target of miRNA-24-3p in GC cells. a The potential miR-24-3p binding sequence of Prdx-6 3′-UTR and the mutated sequence. b miR-24 mimics down-regulated activity of a luciferase reporter containing wild-type Prdx-6 3′-UTR (0.731 ± 0.09), but not the reporter with mutant Prdx-6 3′-UTR (1.182 ± 0.34). Student’s t test with SPSS (version 11.0, IBM, USA) showed a *P = 0.015
miR-24-3p Expression Is Significantly Decreased in Response to H. pylori Infection
To explore the potential role of miR-24-3p in carcinogenesis related to H. pylori infection, the miR-24-3p expression level was measured in a tissue microarray. Compared with the negative controls, ISH revealed that miR-24-3p levels were reduced by 19.6 % in H. pylori-infected tissue specimens. It was also found that miR-24-3p was markedly down-regulated in CSG (50 %) relative to matched H. pylori-non-infected tissues (95.7 %). Compared with the GC H. Pylori-infected tissues, the level of miRNA-24-3p expression was decreased in CAG, IM, and Dys H. Pylori-infected tissues (Table 1).These results suggest that H. pylori infection might contribute to a reduction in miR-24-3p expression in vivo and miR-24-3p levels cannot reduce H. pylori-infected tissue specimens in GC.
miR-24-3p Participates in the Development and Progression of H. pylori-Related Gastric Mucosal Lesions Through Negative Regulation of Prdx-6
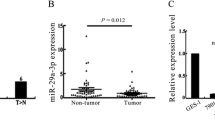
In order to detect Prdx-6 and miR-24-3p expression, the same tissue microarray as well as IHC and in situ hybridization was used. As shown in Fig. 3a, b and Tables 2 and 3, the attenuation of miR-24-3p expression was associated with the development of more severe gastric precancerous mucosa lesions, and the expression of miR-24-3p in gastric tumors was higher than in the adjacent normal mucous tissue as well as in CAG, IM, and Dys. Contrary to these findings, Prdx-6 expression displayed the opposite tendency when examined in the same tissue (Fig. 3c, d; Tables 4, 5). As miR-24-3p negatively regulated Prdx-6 as a direct regulator, it can be further concluded that miR-24-3p as a regulator to target Prdx-6 regulates the development and progression of H. pylori-related gastric mucosal lesions.
miR-24-3p participates in the development and progression of H. pylori-related gastric mucosal lesions through negative regulation of Prdx-6. a Attenuation of miR-24-3p expression was associated with the development of more severe gastric precancerous mucosal lesions, and the expression of miR-24-3p in GC was higher than in CAG, IM, and Dys. b Expression of miR-24-3p in gastric tumors was higher than in the adjacent normal tissue. c, d The expression of Prdx-6 displayed the opposite tendency to miR-24-3p expression in the same tissue specimens
miR-24-3p Level Is Decreased in the Patients with Lymph Node and Liver Metastases
The correlation of miR-24-3p with clinicopathological characteristics was examined in 31 GC patients (Table 6). There was no correlation between miR-24-3p expression and patients’ age, gender, degree of differentiation or gastric carcinoma TNM staging. However, the miR-24-3p level was significantly decreased in the patients who had lymph node [Fisher’s exact test (1-tail)] and liver metastases [Fisher’s exact test (1-tail)]. Since miR-24-3p expression was lowest in N87 cells, which are derived from lymph node metastasis, this cell line was chosen for further study.
miR-24-3p Suppresses Proliferation, Migration, and Invasion and Promotes Apoptosis in N87 Cells by Targeting Prdx-6 mRNA
In order to evaluate the role of miR-24-3p in the N87 cell line, experiments were divided into five groups: Group 1, N87 cells alone; Group 2, N87 cells stably transfected with an empty vector; Group 3, N87 cells stably transfected with miR-24-3p; Group 4, N87 cells transiently transfected with Prdx-6 expression vectors overexpressing empty vector; and Group 5, N87 cells transiently transfected with Prdx-6 expression vectors overexpressing miR-24-3p. The expression of miR-24-3p and Prdx-6 was detected by real-time PCR and Western blot, respectively (Fig. 4a), and the effect of Prdx-6 on miR-24-3p-mediated cell proliferation, apoptosis, migration, and invasion was investigated. N87 cells that had been stably transfected with miR-24-3p (Group 3) demonstrated a lower capacity of proliferation, migration, and invasion and a higher capacity of apoptosis compared with cells that had been stably transfected with the miRNA control (Group 2). These results indicate that miR-24-3p suppresses GC cell proliferation, migration, and invasion and promotes apoptosis. As compared with the stably overexpressed miR-24-3p N87 cells (Group 3), the proliferation, apoptosis, migration, and invasion ability of N87 cells transiently transfected with Prdx-6 expression vectors overexpressing miR-24-3p (Group 5) were partially restored (Fig. 4a–f). These results indicate that miR-24-3p overexpression mediates the suppression of proliferation, migration, and invasion and promotes apoptosis by inhibiting Prdx-6 expression.
miR-24-3p suppresses proliferation, migration, and invasion and promotes apoptosis of N87 cells by targeting Prdx-6. a miR-24-3p was overexpressed in Group 3 and Group 5, Prdx-6 expression in Group 5 was equal to Group 2, and Prdx-6 expression was lowest in Group 3. b Cell proliferation was analyzed by MTT assay in N87 cells. Growth curve is shown for each group at 0, 24, 48, 72, and 96 h. c Representative micrographs and histograms depicting apoptosis of N87 cells in each group after 24 and 48 h. d, e Representative fields of N87 cells in migration and invasion assays, respectively. Gastric cancer cell proliferation, apoptosis, migration, and invasion are restored after Prdx-6 restoration. Average number of cells per field from three independent experiments is presented as data in histograms. Data are presented as mean ± SD or percentage, and means and SD are shown in the bottom of the figure, and Student’s t test with SPSS (version 11.0, IBM, USA) showed a *P < 0.05
Discussion
At present, more than 1900 miRNAs have been identified in humans and they regulate the expression of about 60 % genes in mammals [10]. miRNAs are found to be related to a variety of cellular events, including proliferation, apoptosis. Moreover, miRNAs might be involved in the physiological and pathological processes that contribute to the progress of precancerous lesions to cancer [11–13]. Mortality of GC patients ranks second in all malignancies [14]. Many miRNAs are closely associated with GC, but knowledge of their abnormal expression and potential role in GC is inadequate. Eight miRNAs (miR1274a, miR196b, miR4298, miR181c, miR23a, miR27a, and miR-24-2) were reported to be highly expressed in GC tissue [9]. Combining this result with bioinformatics analysis on Prdx-6, which has been intensively investigated in our previous study [8], we selected miR-24-3p for further study. To explore the negative regulation between miR-24-3p and Prdx-6, the expression of miR-24-3p and Prdx-6 in GES-1 and GC cell lines was examined; then, miR-24-3p mimics, miR-24-3p inhibitor, and negative control were transiently transfected into these cell lines and a negative association between miR-24-3p and Prdx-6 expression was identified in all GC cell lines, except SGC7901. In addition, it was confirmed by a luciferase reporter assay that miR-24-3p directly targets the binding sites in the 3′-UTR of Prdx-6 and represses Prdx-6 protein expression; Prdx-6 gene is one of the direct targets of miR-24-3p. Although a negative association between miR-24-3p and Prdx-6 expression levels was observed in the liver metastasis cell of gastric adenocarcinoma SGC7901, overexpressing miR-24-3p did not decrease the expression of Prdx-6. Since miRNA can regulate several target genes and several miRNAs can coordinate regulation of the same gene, further study is needed to uncover the underlying mechanism in SGC7901.
GC is the final stage of H. pylori-related chronic gastric mucosal lesions, which develops from normal mucosa to CSG, CAG, IM, Dys, and finally to GC [15]. H. Pylori-induced oxidative DNA damage occurs in all phases of this multistep process. In this study, the expression of miR-24-3p was found to decrease along with the progression of precancerous gastric mucosa lesions and miR-24-3p was found to play a part in H. pylori-related gastric mucosal lesions by negative regulation of Prdx-6. These results demonstrate that expression of the Prdx-6 gene is promoted by a decreased expression of miR-24-3p, resulting in reduced ROS-induced gastric mucosal injury and prevention of lesion progression. Furthermore, our data provide evidence of higher miR-24-3p expression level in GC tissues compared to corresponding nontumor normal tissues and miR-24-3p levels cannot reduce H. pylori-infected tissue specimens in GC. The reason for this phenomenon is that with progression of gastric mucosal lesions to GC, ROS was overproduced and miR-24-3p expression was no longer decreased, causing imbalance of the oxidative–antioxidant system. The imbalanced system, in turn, creates feedback that leads to an increased miR-24-3p expression and decreased Prdx-6 expression.
The 5-year survival rate of early diagnosed GC patients is higher than 90 %, while it is only 30-40 % for advanced GC [16, 17]. Our data suggest that miR-24-3p level is dynamically changed in the development and progression of gastric mucosal lesion and related to GC lymph node metastasis, indicating that miR-24-3p can be potentially used for early diagnosis and as a tumor molecular marker to assess the stage of gastric mucosal lesions and lymph node and liver metastasis. There are several limitations in our study. First, the number of samples and cell lines analyzed and validated was not enough. Second, the follow-up data were short, and we cannot get the exact conclusion that miR-24-3p is related to the prognosis.
Conclusions
The 5-year survival rate of early diagnosed GC patients is higher than 90 %, while it is only 30–40 % for advanced GC [16, 17]. Our data suggest that miR-24-3p level is dynamically changed in the development and progression of gastric mucosal lesion and related to GC lymph node metastasis, indicating that miR-24-3p can be potentially used for early diagnosis and as a tumor molecular marker to assess the stage of gastric mucosal lesions and lymph node and liver metastasis.
Abbreviations
- miRNAs:
-
MicroRNAs
- GC:
-
Gastric cancer
- CSG:
-
Chronic superficial gastritis
- Hp:
-
H. pylori
- CAG:
-
Chronic atrophic gastritis
- UTR:
-
Untranslated region
- Prdxs:
-
Peroxiredoxins
- Cys:
-
Cysteine
- CNVs:
-
Copy number variations
- IM:
-
Intestinal metaplasia
- Dys:
-
Dysplasia
- FBS:
-
Fetal bovine serum
- RT-qPCR:
-
Quantitative reverse transcription PCR
- MTT:
-
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- DMSO:
-
Dimethyl sulfoxide
- ICH:
-
Immunohistochemistry
- ISH:
-
In situ hybridization
- SD:
-
Standard deviation
- GES-1:
-
Gastric mucosal epithelial cell
References
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297.
Amiel J, de Pontual L, Henrion-Caude A. miRNA, development and disease. Adv Genet. 2012;80:1–36.
Conrad R, Barrier M, Ford LP. Role of miRNA and miRNA processing factors in development and disease. Birth Defects Res C Embryo Today. 2006;78:107–117.
Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108.
Ishiguro H, Kimura M, Takeyama H. Role of microRNAs in gastric cancer. World J Gastroenterol. 2014;20:5694–5699.
Greenberg JT, Demple B. Overproduction of peroxide-scavenging enzymes in Escherichia coli suppresses spontaneous mutagenesis and sensitivity to redox-cycling agents in oxyR-mutants. EMBO J. 1988;7:2611–2617.
Ambruso DR. Peroxiredoxin-6 and NADPH oxidase activity. Methods Enzymol. 2013;527:145–167.
Guo HW, Lu YY, Zhu YL, Yang GB. Expression of peroxiredoxin 6 in gastric cancer and its clinical significance. Zhonghua Yi Xue Za Zhi. 2012;92:2433–2435.
An J, Pan Y, Yan Z, et al. MiR-23a in amplified 19p13.13 loci targets metallothionein 2A and promotes growth in gastric cancer cells. J Cell Biochem. 2013;114:2160–2169.
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874.
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531.
Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184.
Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917.
Choi H, Chang JW, Jung YK. Peroxiredoxin 6 interferes with TRAIL-induced death-inducing signaling complex formation by binding to death effector domain caspase. Cell Death Differ. 2011;18:405–414.
Ajani JA, Bentrem DJ, Besh S, et al. Gastric cancer, version 2.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2013;11:531–546.
Acknowledgments
This work is supported by Beijing Natural Science Foundation (Grant: 7132207).
Authors’ contributions
Qian Li carried out the molecular genetic studies, Qian Li, Nina Wang participated in the sequence alignment and drafted the manuscript. Guibin Yang, Qian Li, Nina Wang participated in the design of the study and performed the statistical analysis. Hong Wei, Chao Li conceived of the study and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
None.
Ethical statements
Written informed consent from the donor was obtained for the use of this sample in research. The study was approved by the Ethics Committee of Aerospace Center Hospital (20140627YJS-01).
Additional information
Jing Wu and Guibin Yang are corresponding authors.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Li, Q., Wang, N., Wei, H. et al. miR-24-3p Regulates Progression of Gastric Mucosal Lesions and Suppresses Proliferation and Invasiveness of N87 Via Peroxiredoxin 6. Dig Dis Sci 61, 3486–3497 (2016). https://doi.org/10.1007/s10620-016-4309-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-016-4309-9