Abstract
Federated learning (FL) is emerging as a privacy-aware alternative to classical cloud-based machine learning. In FL, the sensitive data remains in data silos and only aggregated parameters are exchanged. Hospitals and research institutions which are not willing to share their data can join a federated study without breaching confidentiality. In addition to the extreme sensitivity of biomedical data, the high dimensionality poses a challenge in the context of federated genome-wide association studies (GWAS). In this article, we present a federated singular value decomposition algorithm, suitable for the privacy-related and computational requirements of GWAS. Notably, the algorithm has a transmission cost independent of the number of samples and is only weakly dependent on the number of features, because the singular vectors corresponding to the samples are never exchanged and the vectors associated with the features are only transmitted to an aggregator for a fixed number of iterations. Although motivated by GWAS, the algorithm is generically applicable for both horizontally and vertically partitioned data.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
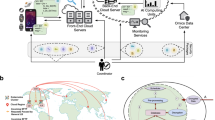
Federated learning (FL) has recently gained attention as a privacy-aware alternative to centralized computation. Unlike in centralized machine learning (ML), where the data is consolidated at a central server and a model is calculated on the combined data, in FL, the data remains with the data owners (Mothukuri et al. 2021). Instead of the data, only model parameters are sent to the other parties, such that the the local models are aggregated into a global model (see Fig. 1 for a schematic visualization of FL).
Schematic comparison of traditional cloud-based approaches (left) and FL (right). In the cloud-based approach, data contributors send their private data, represented by padlocks, to a central server, where the model is computed. Thereby, data contributors lose agency over their data. In FL, the raw data remains with the data owners and only parameters are exchanged
FL is subdivided in cross-device and cross-silo FL. Cross-device FL assumes a high number of devices (e.g., mobile phones or sensors) with limited compute power to be connected in a dynamic fashion, meaning that clients are expected to join and drop out during the learning process. Cross-silo FL has a lower number of participants, which hold a larger amount of data, have higher compute power, and are connected in a more static fashion. Clients are not expected to join and drop out during the learning process randomly (Kairouz et al. 2021). FL is distinct from distributed computing, where the data resides in one location but the operations are distributed on several worker nodes.
An attractive application case for cross-silo FL are genome-wide association studies (GWAS), which investigate the relationship of genetic variation with phenotypic traits on large cohorts (Visscher et al. 2017; Tam et al. 2019). Since genetic data is extremely sensitive, data holders cannot make it publicly available. The practical feasibility of using FL for GWAS has been demonstrated recently (Nasirigerdeh et al. 2020; Cho et al. 2018).
Since GWAS are often carried out on populations of mixed ancestry, cryptic population confounders should be controlled for before associating the genetic variants to the phenotypic trait of interest. These population confounders represent hidden substructures in the cohort which are difficult to control for using categorical covariates (e.g., genetic ancestry). The standard way for achieving this is to compute the leading eigenvectors of the sample covariance matrix via principal component analysis (PCA) and to include these eigenvectors as confounding variables to the models used for the association tests (Price et al. 2006; Galinsky et al. 2016).
For federated GWAS, a PCA algorithm for vertically partitioned data is required to compute the eigenvectors (see Sect. 2 for a detailed explanation). Although a few such algorithms are available (Kargupta et al. 2001; Qi et al. 2003; Guo et al. 2012; Wu et al. 2018), none of them is suitable for federated GWAS. More precisely, most of the algorithms reviewed by Wu et al. (2018) focus on finding a consensus in mesh networks, which is very dissimilar to the setup used in GWAS, where relatively few data holders collaborate in a static setting. The algorithms presented by Kargupta et al. (2001) and Qi et al. (2003) rely on estimating a proxy covariance matrix and hence do not scale to large GWAS datasets, which often contain genetic variation data for hundreds of thousands of individuals. One of the few covariance-free PCA algorithm suitable for an architecture with a limited number of peers has been presented by Guo et al. (2012). However, this algorithm broadcasts the complete first \(k-1\) sample eigenvectors to the aggregator, which constitutes a privacy leakage that should be avoided in federated GWAS (Nasirigerdeh et al. 2021). Cho et al. (2018) present a secure multiparty protocol for GWAS which uses PCA. The protocol includes three external parties and potential physical shipping of data. The setup is fundamentally different: The data holders are individuals who only have access to one record. They create secret shares which are processed by two computing parties.
In previous algorithms, including algorithms designed for horizontally partitioned data as described by Balcan et al. (2014), the exchanged parameters scale with the number of genetic variants (features) in the dataset as the feature eigenvectors are exchanged. At the scale of GWAS with several million genetic variants, this is another challenge for the existing algorithms. Furthermore, due to the iterative nature of the algorithm, the process is prone to information leakage, a problem previously not investigated. More precisely, the feature eigenvector updates exchanged during the learning process can be used to compute the feature covariance matrix, given a sufficient number of iterations. In terms of disclosed information, this makes the algorithm equivalent to algorithms exchanging the entire covariance matrix. The feature covariance matrix is a summary statistic over all samples, but due to its size contains a high amount of information and can be used to generate realistically looking samples. Therefore, the communication of the entire feature eigenvectors should also be avoided as far as possible.
Extrapolating from the shortcomings of existing approaches, we can state that, for federated GWAS, a PCA algorithm for vertically partitioned data is required that combines the following properties:
-
The algorithm should be suitable for a cross-silo FL architecture with few static participants.
-
The algorithm should not rely on computing or approximating the covariance matrix.
-
The algorithm should be communication-efficient.
-
The algorithm should avoid the communication of the sample eigenvectors and reduce the communication of the feature eigenvectors.
In this paper, we present the first algorithm that combines all of these desirable properties and can hence be used for federated GWAS (and all other applications where these properties are required). We prove that our algorithm is equivalent to centralized vertical subspace iteration (Halko et al. 2010)—a state-of-the-art centralized, covariance-free SVD algorithm—and therefore generically applicable to any kind of data. Thereby, we show that the notion of “horizontally” and “vertically” partitioned data are irrelevant for SVD. Furthermore, we apply two strategies to make the algorithm more communication-efficient, both in terms of communication rounds and transmitted data volume. More specifically, we employ approximate PCA (Balcan et al. 2014) and randomized PCA (Halko et al. 2010). We show in an empirical evaluation that the eigenvectors computed by our approaches converge to the centrally computed eigenvectors after sufficiently many iterations. In sum, the article contains the following main contributions:
-
We present a federated PCA algorithm for vertically partitioned data which meets the requirements that apply in federated GWAS settings.
-
We prove that our algorithm is equivalent to centralized power iteration and show that it exhibits an excellent convergence behavior in practice.
-
Our algorithm is generically applicable for federated SVD on both “horizontally” and “vertically” partitioned data.
This article is an extended and consolidated version of a previous conference publication (Hartebrodt et al. 2021) with the following additional contributions: a demonstration how iterative leakage can pose a problem for federated power iteration; a further reduction in transmission cost, and increase in privacy, due to the use of randomized PCA; a data-dependent speedup due to the use of approximate PCA. The remainder of this paper is organized as follows: In Sect. 2, we introduce concepts and notations that are used throughout the paper and discuss related work. In Sect. 3, we describe the proposed algorithms. We then describe how to extract the covariance matrix from the updates in Sect. 4. In Sect. 5, we report the results of the experiments. Section 6 concludes the paper.
2 Preliminaries and related work
2.1 Federated learning, employed data model, and notations
Typically, a cross-silo architecture with a limited number of peers is used in biomedical federated solutions (Steed and Fradinho Duarte de Oliveira 2010; Nasirigerdeh et al. 2020), with the data holders acting as clients. The datasets at the client sites will be called local datasets. The parameters or models learned using this data will be called local parameters or local models, while the final aggregated model will be called global model. The optimal result of the global model is achieved when it equals the result of the conventional model calculated on all data, which we call the centralized model.
In federated settings, the data can be distributed in several ways. Either the clients observe a full set of variables for a subset of the samples (horizontal partitioning) or they have a partial set of variables for all samples (vertical partitioning) (Rodríguez et al. 2017; Wu et al. 2018). In this paper, we assume that we are given a global data matrix \({\textbf{A}} \in \mathbb {R}^{m\times n}\), where m is the number of features (genetic variants, in the context of GWAS) and n is the overall number of samples. However, this global data matrix is never materialized. Instead, the data is split across S local sites as \({\textbf{A}} =[{\textbf{A}} ^1\dots {\textbf{A}} ^s\dots {\textbf{A}} ^S]\), where \({\textbf{A}} ^s\in \mathbb {R}^{m\times n^s}\) and \(n^s\) denotes the number of samples available at site s. From a semantic point of view, the partitioning is hence horizontal, since the samples are distributed over the local sites. However, from a technical point of view, the partitioning is vertical, since the samples correspond to the columns of \({\textbf{A}}\). The reason for this rather unintuitive setup is that, when using PCA for GWAS, samples are treated as features, as detailed in the following paragraphs.
Table 1 provides an overview of notations which are used throughout the paper.
2.2 Principal component analysis and singular value decomposition
Given a data matrix \({\textbf{A}} \in \mathbb {R}^{m \times n}\), the PCA is the decomposition of the covariance matrix \({\textbf{M}} ={\textbf{A}} ^\top {\textbf{A}} \in \mathbb {R}^{n \times n}\) into \({\textbf{M}} = {\textbf{V}} \varvec{\Sigma } {\textbf{V}} ^ \top\). \(\varvec{\Sigma }\in \mathbb {R}^{n\times n}\) is a diagonal matrix containing the eigenvalues \((\sigma _{i})_{i=1}^n\) of \({\textbf{M}}\) in non-increasing order, and \({\textbf{V}} \in \mathbb {R}^{n \times n}\) is the corresponding matrix of eigenvectors (Joliffe 2002). Singular value decomposition is closely related to PCA and an extension of PCA to non-square matrices (in fact, many PCA algorithms call SVD solvers to do the actual computation). Given a data matrix \({\textbf{A}} \in \mathbb {R}^{m \times n}\), the SVD is its decomposition into \({\textbf{A}} = {\textbf{U}} \varvec{\Sigma } {\textbf{V}} ^ \top\). The matrices \({\textbf{U}}\) and \({\textbf{V}}\) are the left and right singular vector matrices. Usually, one is only interested in the top k eigenvalues and corresponding eigenvectors. Since k is arbitrary but fixed throughout this paper, we let \({\textbf{G}} \in \mathbb {R}^{n\times k}\) and \({\textbf{H}} \in \mathbb {R}^{m\times k}\) denote these first k eigenvectors (i.e., , \({\textbf{G}}\) corresponds to the first k columns of \({\textbf{V}}\)). \({\textbf{G}}\) is typically used to obtain a low-dimensional representation \({\textbf{A}} ^\top \mapsto {\textbf{A}} ^\top {\textbf{H}} \in \mathbb {R}^{n\times k}\) of the data matrix \({\textbf{A}}\), which can then be used for downstream data analysis tasks. This, however, is not the way PCA is used in GWAS, as we will explain next.
2.3 Genome-wide association studies
The genome stores hereditary information that controls the phenotype of an individual in interplay with the environment. The genetic information stored in the DNA is encoded as a sequence of bases (A, T, C, G). Positions in this sequence are called loci. If we observe two or more possible bases at a specific locus in a population, we call this locus a single-nucleotide polymorphism (SNP). The predominant base in a population is called the major allele; bases at lower frequency are called minor alleles (Tam et al. 2019).
GWAS seek to identify SNPs that are linked to a specific phenotype (Visscher et al. 2017; Tam et al. 2019). Phenotypes of interest can for example be the presence or absence of diseases, or quantitative traits such as height or body mass index. The SNPs for a large cohort of individuals are tested for association with the trait of interest. Typically, simple models such as linear or logistic regression are used for this (Visscher et al. 2017; Nasirigerdeh et al. 2020). The input to a GWAS is an n-dimensional phenotype vector \({\textbf{y}}\), a matrix of SNPs \({\textbf{A}} \in \mathbb {R}^{m\times n}\), and confounding factors such as age or sex, given as column vectors \({\textbf{x}} _r\in \mathbb {R}^{n}\). The SNPs are encoded as categorical values between 0 and 2, representing the number of minor alleles observed in the individual at the respective position. Each SNP \(l\in [m]\) is tested in an individual association test
where \({\textbf{A}} _{l,\bullet }\) denotes the lth row of \({\textbf{A}}\).
2.4 Principal component analysis for genome-wide association studies
Confounding factors such as ancestry and population substructure can alter the outcome of an association test and can create false hits if not properly controlled for (Tam et al. 2019). PCA has emerged as a popular strategy to infer population substructure and a protocol based on secure multiparty computation (SMPC) has been presented by Cho et al. (2018). More precisely, the first k (usually \(k=10\)) eigenvectors \({\textbf{G}} =[{\textbf{g}} _1\dots {\textbf{g}} _k]\in \mathbb {R}^{n\times k}\) of the sample covariance matrix \({\textbf{A}} ^\top {\textbf{A}}\) are included into the association test as covariates (Galinsky et al. 2016; Price et al. 2006):
In federated GWAS, each local site s needs to have access only to the partial eigenvector matrix \({\textbf{G}} ^s\) corresponding to the locally available samples. Consequently, computing the complete eigenvector matrix \({\textbf{G}}\) at the aggregator and/or sharing \({\textbf{G}} ^s\) with other local sites \(s^\prime\) should be avoided to reduce the possibility of information leakage. This is especially important because Nasirigerdeh et al. (2021) have shown that, if \({\textbf{G}}\) is available at the aggregator in a federated GWAS pipeline, the aggregator can in principle reconstruct the raw GWAS data \({\textbf{A}} _{l,\bullet }\) for SNP l. Federated PCA algorithms that are suitable for GWAS hence have to respect the following constraint:
Constraint 1
In a GWAS-suitable federated PCA algorithm, the aggregator does not have access to the complete eigenvector matrix \({\textbf{G}}\) and each site s has access only to its share \({\textbf{G}} ^s\) of \({\textbf{G}}\).
The PCA in GWAS is usually performed on only a subsample of the SNPs, but there is no consensus as to how many SNPs should be used. Some PCA-based stratification methods rely on a small set of ancestry-informative markers (Li et al. 2016), while others employ over 100000 SNPs (Gauch et al. 2019).
Note that PCA for GWAS is conceptually different from “regular” PCA for feature reduction (see Fig. 2). For feature reduction, we would decompose the \(m \times m\) SNP by SNP covariance matrix and compute a set of “meta-SNPs” for each sample. This is not what is required for GWAS. Instead, the \(n \times n\) sample by sample covariance matrix \({\textbf{A}} ^\top {\textbf{A}}\) is decomposed. In our federated setting, where \({\textbf{A}}\) is vertically distributed across local sites \(s\in [S]\), \({\textbf{A}} ^\top {\textbf{A}}\) looks as follows (recall that, unlike in regular PCA, columns correspond to samples and rows to features):
It is clear that \({\textbf{A}} ^\top {\textbf{A}}\) cannot be computed directly without sharing patient-level data. Moreover, with a growing number of samples, this matrix can become very large and computing it becomes infeasible. For instance, the UK Biobank—a large cohort frequently used for GWAS—contains more than 4 million SNPs for more than 500000 individuals. Following directly from the definition of PCA, an exact computation of the covariance matrix would furthermore violate constraint 1. These considerations lead to the second constraint for federated PCA algorithms suitable for GWAS:
Constraint 2
A GWAS-suitable federated PCA algorithm must work on vertically partitioned data and does not rely on computing or approximating the covariance matrix.
2.5 Gram–Schmidt orthonormalization
The Gram–Schmidt algorithm transforms a set of linearly independent vectors into a set of mutually orthogonal vectors. Given a matrix \({\textbf{V}} =[{\textbf{v}} _1\dots {\textbf{v}} _k]\in \mathbb {R}^{r\times k}\) of k linearly independent column vectors, a matrix \({\textbf{U}} =[{\textbf{u}} _1\dots {\textbf{u}} _k]\in \mathbb {R}^{r\times k}\) of orthogonal column vectors with the same span can be computed as
where \(r_{i,j}={\textbf{u}} _j^\top {\textbf{v}} _i/n_j\) with \(n_j={\textbf{u}} _j^\top {\textbf{u}} _j\).
The vectors can then be scaled to unit Euclidean norm as \({\textbf{u}} _i\mapsto (1/\sqrt{n_i})\cdot {\textbf{u}} _i\) to achieve a set of orthonormal vectors. In the context of PCA, this can be used to ensure orthonormality of the candidate eigenvectors in iterative procedures, which otherwise suffer from numerical instability in practice (Guo et al. 2012).
2.6 Centralized, iterative, covariance-free principal component analysis
While classical PCA algorithms rely on computing the covariance matrix \({\textbf{A}} ^\top {\textbf{A}}\) (Joliffe 2002), there are several covariance-free approaches to iteratively approximate the top k eigenvalues and eigenvectors (Saad 2011). Algorithm 1 summarizes the centralized, iterative, covariance-free PCA algorithm suggested by Halko et al. (2010), which will serve as starting point for our federated approach. First, an initial eigenvector matrix is sampled randomly and is then orthonormalized (Lines 1−2). In every iteration i, improved candidate eigenvectors \({\textbf{G}} _i\) of \({\textbf{A}} ^\top {\textbf{A}}\) are computed (Lines 5–8). Once a suitable termination criterion is met (e.g., convergence, maximal number of iterations, time limit, etc.), the last candidate eigenvectors are returned (Line 10).

To update the candidate eigenvector matrices \({\textbf{G}} _i={\textbf{A}} ^\top {\textbf{H}} _i={\textbf{A}} ^\top {\textbf{A}} {\textbf{G}} _{i-1}\in \mathbb {R}^{n\times k}\) of \({\textbf{A}} ^\top {\textbf{A}}\), the algorithm also computes candidate eigenvector matrices \({\textbf{H}} _i={\textbf{A}} {\textbf{G}} _{i-1} = {\textbf{A}} {\textbf{A}} ^\top {\textbf{H}} _{i-1}\in \mathbb {R}^{m\times k}\) of \({\textbf{A}} {\textbf{A}} ^\top\). Since \({\textbf{A}} {\textbf{A}} ^\top\) corresponds to the “classical” feature by feature covariance matrix, and \({\textbf{A}} ^\top {\textbf{A}}\) to the sample covariance matrix, the algorithm computes left and right singular vectors at the same time. This means, the present algorithm is actually an SVD algorithm. In this article, we will sometimes refer to left singular vectors as feature eigenvectors and right singular vectors as sample eigenvectors.
2.7 Federated principal component analysis for vertically partitioned data
Only few algorithms are designed to perform federated computation of PCA on vertically partitioned data sets (Guo et al. 2012; Kargupta et al. 2001; Qi et al. 2003; Wu et al. 2018). However, none of them is suitable for the GWAS use-case considered in this paper: The algorithms reviewed by Wu et al. (2018) are specialised for distributed sensor networks and use gossip protocols and peer-to-peer communication. Therefore, they are not suited for the intended FL architecture in the medical setting. The algorithms presented by Kargupta et al. (2001) and Qi et al. (2003) rely on estimating a proxy covariance matrix and consequently do not meet constraint 2 introduced above. Unlike these approaches, the algorithm proposed by Guo et al. (2012) is covariance-free and suitable for the intended cross-silo architecture. However, it broadcasts the eigenvectors to all sites in violation of constraint 1.
2.8 Federated matrix orthonormalization
Matrix orthonormalization is a frequently used technique in many applications, including the solution of linear systems of equations and singular value decomposition. There are three main approaches: Householder reflection, Givens rotation, and the Gram–Schmidt algorithm. In distributed memory systems and grid architectures, tiled Householder reflection is a popular approach (Hadri et al. 2010; Hoemmen 2011). However, those algorithms are often highly specialized to the compute system and rely on shared disk storage. For distributed sensor networks, Gram–Schmidt procedures relying on push-sum have been proposed (Sluciak et al. 2016; Straková et al. 2012). However, these methods rely on gossip protocols and are not appropriate for the network architecture in this article. Consequently, no federated orthonormalization algorithm suitable for our setup is available. In Sect. 3.1, we present our own version of a federated orthonormalization algorithm fulfilling all constraints and subsequently utilize it as a subroutine in our federated PCA algorithm.
2.9 Federated principal component analysis for horizontally partitioned data
Previously, federated PCA algorithms have been described for horizontal data partitioning. In a previous article (Hartebrodt and Röttger 2022), we discuss potential algorithms for this task, summarize their advantages and drawbacks in the context of biomedical applications, and provide practical guidelines for users from the biomedical sciences. There are “single-round” approaches, where the eigenvectors are computed locally and sent to the aggregator (Balcan et al. 2014). At the aggregator, a global subspace is approximated from the local eigenspaces. The higher the number of transmitted intermediate dimensions, the better the global subspace approximation. In these algorithms, the solution quality hence depends on the number of transmitted dimensions. This algorithm is a more memory efficient version of the naïve algorithm (Liu et al. 2020), where the entire covariance matrix is processed by the aggregator. Since only the top k left singular values are transmitted, this algorithm fulfills constraint 2. Furthermore, iterative schemes have been proposed, where locally computed eigenvectors are sent to the aggregator, which performs an aggregation step and sends the obtained candidate subspace back to the clients (Balcan et al. 2016; Chen et al. 2021; Imtiaz and Sarwate 2018). The candidate subspace is then refined iteratively. Furthermore, there are several schemes for specific applications such as streaming (Sanchez-Fernandez et al. 2015; Grammenos et al. 2020). These approaches assume that an approximation of the entire eigenvectors is possible at the clients, or that the global covariance matrix can be approximated. As we have discussed above, these assumptions do not hold in the intended GWAS use case.
2.10 Randomized principal component analysis
In the context of GWAS, a randomized PCA algorithm (Halko et al. 2010; Galinsky et al. 2016) is popular as it speeds-up the computation compared to traditional algorithms. Here, we briefly present the version proposed by Galinsky et al. (2016). The algorithm starts with \(I^\prime\) iterations of subspace iteration on the full-dimensional data matrix, resulting in feature eigenvector matrices \({\textbf{H}} _i\) for all iterations \(i\in \{1,\ldots ,I^\prime \}\). Next, the data is projected on the concatenation of all \({\textbf{H}} _i\), forming approximate principal components which approximate the data matrices. Then, subspace iteration is performed on these proxy data matrices. In practice, \(I^\prime =10\) iterations are sufficient. This reduces the dimensionality of the data from m to \(k\cdot I^\prime\). In Sect. 3.3, we will present a fully federated version of this algorithm in detail. Note that this is a randomized approach, because subspace iteration on the full-dimensional data is initialized randomly. Since it is interrupted before convergence after \(I^\prime\) iterations, the feature eigenvector matrices \({\textbf{H}} _i\) inherit this randomness.
2.11 Privacy-aware singular value decomposition
In the context of privacy-aware ML, several solutions have been proposed to allow the private computation of PCA and SVD. Differential privacy (DP) has been employed by multiple authors who suggest to privately construct a covariance matrix (Balcan et al. 2016; Chaudhuri et al. 2013; Imtiaz and Sarwate 2018; Imtiaz et al. 2019; Wang and Chang 2019). Similarly, the secure aggregation of the covariance matrix using either secure multiparty computation (SMPC) or homomorphic encryption (HE) has been suggested (Al-Rubaie et al. 2017; Liu et al. 2020; Pathak and Raj 2011). Hardt and Price (2013) proposes a differentially private power method. Asi and Duchi (2020) and Gonen et al. (2018) suggest to use output perturbation, assuming an SVD has been computed. Recently, Grammenos et al. (2020) have suggested a federated streaming PCA scheme, allowing the addition of noise at the nodes with favorable noise variance. However, Chai et al. (2021) deem the addition of noise prohibitive. HE-based methods incur a high runtime overhead (Chai et al. 2021), not yet suitable for large-scale computations. Cho et al. (2018) rely on sharding the data to two non-colluding servers. Froelicher et al. (2023) recently proposed a hybrid approach, relying on SMPC and HE. Their approach significantly improves upon the runtime of previous HE-based methods but still incurs a high runtime overhead. In cross-device FL, other solutions using third party servers have been suggested, e.g., by Liu and Tang (2019); Cho et al. (2018). However, Liu and Tang (2019) assume that the computation of the covariance matrix is computationally feasible.
3 Algorithms
In this section, we present a federated SVD algorithm, which is designed for a cross-silo FL setup, meets the requirements of constraint 1 and constraint 2, and is hence suitable for federated GWAS. Our base algorithm comes in two variants—with and without orthonormalization of the candidate right singular vectors of \({\textbf{A}}\). In Sect. 3.1, we present a federated Gram–Schmidt algorithm, which can be used as a subroutine in our federated SVD algorithm to ensure that right singular vectors of \({\textbf{A}}\) remain at the local sites. We prove that our federated Gram–Schmidt algorithm is equivalent to the centralized counterpart. In Sect. 3.2, we describe the federated SVD algorithm and prove that the version with orthonormalization is equivalent to the centralized vertical subspace iteration algorithm by Halko et al. (2010), which we have summarized in Algorithm 1 above. We then show how approximate horizontal PCA can be used to compute approximate principal components for immediate use or as initialization for subspace iteration in Sect. 3.3. In Sect. 3.4, we present randomized federated subspace iteration as a means to reduce the transmission cost in federated SVD. In addition to decreasing the communication cost, the use of the two latter strategies also prevents potential iterative leakage (detailed in Sect. 4). In Sect. 3.5, we analyze the network transmission costs of the proposed algorithms, and Sect. 3.6 provides an overview of possible configurations of our federated SVD algorithm. Note that, for the ease of notation, we describe our algorithm in terms of a star-like architecture. The algorithm can be deployed in a peer-to-peer system without loss of precision, but at the expense of a higher communication overhead.
3.1 Federated Gram–Schmidt algorithm
First, we describe federated Gram–Schmidt orthonormalization for vertically partitioned column vectors. Previous federated PCA algorithms require the complete eigenvectors to be known at all sites for the orthonormalization procedure. The naïve way of orthonormalizing the eigenvector matrices in a federated fashion would be to send them to the aggregator which performs the aggregation and then sends the orthonormal matrices back to the clients. However, in this naïve scheme, the transmission cost scales with the number of variables (individuals in GWAS) and all eigenvectors are known to the aggregator.
To address these two problems, we suggest a federated Gram–Schmidt orthonormalization procedure, summarized in Algorithm 2. The algorithm exploits the fact that the computations of the squared norms \(n_i\) and of the residuals \(r_{ij}\) can be decomposed into independent computations of summands \(n_i^s\) and \(r_{ij}^s\) computable at the local sites \(s\in [S]\). The clients compute the local summands and send them to the aggregator, where the squared norm of the first orthogonal vector is computed and sent to the clients (Lines 2–6). Subsequently, the remaining \(k-1\) vectors are orthogonalized. For the ith vector \({\textbf{v}} _i\), the algorithm computes the residuals \(r_{ij}\) w.r.t. all already computed orthogonal vectors \({\textbf{u}} _j\), using the fact that the corresponding squared norms \(n_j\) are already available (Lines 10 to 12). The residuals are aggregated by the central server (Lines 14 to 15). Next, \({\textbf{v}} _i\) is orthogonalized at the clients, the local norms are computed (Lines 18 to 20), and the squared norm of the resulting orthogonal vector \({\textbf{u}} _i\) is computed at the aggregator and sent back to the clients (Line 22). After orthogonalization, all orthogonal vectors are scaled to unit norm at the clients (Lines 25–28). This procedure returns exactly the same result as the centralized algorithm, as shown in Proposition 1 below.

Proposition 1
Centralized and federated Gram–Schmidt orthonormalization are equivalent.
Proof
Let \({\textbf{V}} =[{\textbf{v}} _1\dots {\textbf{v}} _k]\) be the matrix that should be orthonormalized, \({\textbf{v}} _i^s\) be the restriction of the ith columns vector to the samples available at site s, and \({\textbf{u}} _i^s\) be the restriction of the ith orthogonal vector computed by the centralized Gram–Schmidt algorithm before normalization to the samples available at site s. Moreover, let \(n_i\) and \(r_{i,j}\) be the centrally computed norms and residuals, and \({\tilde{n}}_i\), \({\tilde{r}}_{i,j}\), and \({\tilde{{\textbf{u}}}}_i^s\) be the locally computed norms, residuals, and partial orthogonal vectors before normalization. We show by induction on i that \(n_i={\tilde{n}}_i\), \(r_{ij}={\tilde{r}}_{ij}\), and \({\textbf{u}} _i^s={\tilde{{\textbf{u}}}}_i^s\) holds for all \(i\in [k]\) and all \(j\in [i-1]\). This implies the proposition.
For \(i=1\), we have \({\textbf{u}} _1^s={\textbf{v}} _1^s={\tilde{{\textbf{u}}}}_1^s\) and \(n_1={\textbf{u}} _1^\top {\textbf{u}} _1=\sum _{s=1}^S{{\textbf{u}} _1^s}^\top {\textbf{u}} _1^s=\sum _{s=1}^S{{\tilde{{\textbf{u}}}}^{s\top }_1}{\tilde{{\textbf{u}}}}_1^s={\tilde{n}}_1\). For the inductive step, note that \(r_{ij}={{\textbf{u}} _j}^\top {\textbf{v}} _i/n_j=\sum _{s=1}^S{{\textbf{u}} _j^s}^\top {\textbf{v}} _i^s/n_j=\sum _{s=1}^S{\tilde{{\textbf{u}}}}_j^{s\top }{\textbf{v}} _i^s/{\tilde{n}}_j={\tilde{r}}_{ij}\), where the third identity follows from the inductive assumption. Moreover, we have \({\textbf{u}} _{i}^s={\textbf{v}} _{i}^s-\sum _{j=1}^{i-1}r_{ij}\cdot {\textbf{u}} _{j}^s={\textbf{v}} _{i}^s-\sum _{j=1}^{i-1}{\tilde{r}}_{ij}\cdot {\tilde{{\textbf{u}}}}_{j}^s={\tilde{{\textbf{u}}}}_{i}^s\), where the second identity follows from the inductive assumption and the identities \(r_{ij}={\tilde{r}}_{ij}\) established before. We hence obtain \(n_i={\textbf{u}} _i^\top {\textbf{u}} _i=\sum _{s=1}^S{{\textbf{u}} _i^s}^\top {\textbf{u}} _i^s=\sum _{s=1}^S{{\tilde{{\textbf{u}}}}^{s\top }_i}{\tilde{{\textbf{u}}}}_i^s={\tilde{n}}_i\), which completes the proof. \(\square\)
3.2 Federated vertical subspace iteration
Algorithm 3 describes our federated vertical subspace iteration algorithm: Initially, the first partial candidate right singular matrices \({\textbf{G}} _0^s\) of \({\textbf{A}}\) are generated randomly and orthonormalized (Lines 6–9). Inside the main loop, the left singular vectors are updated at the clients, summed up element-wise and orthonormalized at the aggregator, and then sent back to the clients (Lines 14–17). Next, the clients update the partial right singular vectors (Line 21). In the version with orthonormalization, the candidate right singular vectors are now normalized by calling the federated Gram–Schmidt orthonormalization algorithm (Line 24) presented in Sect. 3.1 (Algorithm 2). Note that this algorithm ensures that the partial singular vectors \({\textbf{G}} _i^s\) remain at the local sites. Finally, the full left singular matrices \({\textbf{H}}\) and the orthonormalized partial right singular matrices \({\textbf{G}} ^s\) are returned to the clients (Line 30). In practice, the federated orthonormalization of \({\textbf{G}} _i\) (Line 24) may be omitted to speed up computation. Note, however, that \({\textbf{H}} _i\) is still orthonormalized in every iteration and that the final orthonormalization (Line 28) is required.

Like the original centralized version described in Algorithm 1 above, our algorithm can be run with various termination criteria. In our implementation, we use the convergence criterion
using the angle as a global measure as suggested by Lei et al. (2016), where \({\textbf{1}}_k\) is the k-dimensional vector of ones and t is a small positive number. With this criterion, the algorithm terminates once all right singular vectors of \({\textbf{A}}\) are asymptotically collinear with respect to the eigenvectors of the previous iteration. Other convergence criteria could be used as drop-in replacements.
We now prove that the version of Algorithm 3 with orthonormalization is equivalent to the centralized version described in Algorithm 1. Thus, it inherits its convergence behavior from the centralized version.
Proposition 2
If orthonormalization is used, centralized and federated vertical subspace iteration are equivalent.
Proof
Let \({\textbf{G}} _i\) and \({\textbf{H}} _i\) denote the eigenvector matrices maintained by the centralized algorithm described in Algorithm 1 at the end of the main while-loop, and \({\textbf{G}} _i^s\) be the sub-matrix of \({\textbf{G}} _i\) for the samples available at site s. Moreover, let \({\tilde{{\textbf{H}}}}_i\), \({\tilde{{\textbf{G}}}}_i\), \({\tilde{{\textbf{G}}}}^s_i\), and \({\tilde{{\textbf{H}}}}^s_i\) be the (partial) eigenvector matrices maintained by our federated Algorithm 3 at the end of the main while-loop. We will show by induction on the iterations i that \({\textbf{H}} _i={\tilde{{\textbf{H}}}}_i\) and \({\textbf{G}} _i^s={\tilde{{\textbf{G}}}}_i^s\) for all \(s\in [S]\) holds throughout the algorithm, if the same random seeds are used for initialization.
For \(i=0\), we only have to show \({\textbf{G}} _0^s={\tilde{{\textbf{G}}}}_0^s\). This directly follows from Proposition 1 and our assumption that the same random seeds are used for initialization. For the inductive step, note that, before orthonormalization in Line 17, we have \({\tilde{{\textbf{H}}}}_i=\sum _{s=1}^S{\tilde{{\textbf{H}}}}_i^s=\sum _{s=1}^S{\textbf{A}} ^s{\tilde{{\textbf{G}}}}_{i-1}^s=\sum _{s=1}^S{\textbf{A}} ^s{\textbf{G}} _{i-1}^s={\textbf{A}} {\textbf{G}} _{i-1}={\textbf{H}} _i\), where the third equality follows from the inductive assumption. Because of Proposition 1, this identity continues to hold at the end of the main while-loop.
Similarly, after updating in Line 21 but before orthonormalization, we have \({\tilde{{\textbf{G}}}}^s_i={{\textbf{A}} ^s}^\top {\tilde{{\textbf{H}}}}_i={{\textbf{A}} ^s}^\top {\textbf{H}} _i=({\textbf{A}} ^\top {\textbf{H}} _i)^s={\textbf{G}} _i^s\), where the second equality follows the identity \({\textbf{H}} _i={\tilde{{\textbf{H}}}}_i\) shown above and \(({\textbf{A}} ^\top {\textbf{H}} _i)^s\) denotes the sub-matrix of \({\textbf{A}} ^\top {\textbf{H}} _i\) for the samples available at site s. Again, Proposition 1 ensures that the identity continues to hold after orthonormalization. \(\square\)
The omission of the orthonormalization of \({\textbf{G}} _i\) (Algorithm 3, Line 9 and Line 24) removes provable identity to Algorithm 1. However, other formulations of centralized power iteration exist which directly operate on the covariance matrix (Balcan et al. 2016). In these schemes, instead of splitting the iteration into \({\textbf{H}} _i\) update (Algorithm 1, Line 5) and \({\textbf{G}} _i\) update (Algorithm 1, Line 7), the covariance matrix is computed and \({\textbf{H}} _i\) is updated as \({\textbf{H}} _i={\textbf{A}} {\textbf{A}} ^\top {\textbf{H}} _{i-1}\) at every iteration. Proposition 2 can be formulated and proven analogously for this version.
3.3 Approximate initialization
One major concern of iterative PCA is information leakage through the repeated transmission of the updated eigenvectors. This is presented in more detail in Sect. 4, where we will present an attack which leaks the covariance matrix. Briefly, the consequence of the possible attack is that the number of iterations needs to be strictly limited. Therefore, we suggest to use federated approximate horizontal PCA as an initialization strategy to limit the number of iterations, and thereby prevent the possible leakage of the covariance matrix.
Balcan et al. (2014) presented a memory-efficient version of federated approximate PCA for horizontally partitioned data. We provide a minor modification which allows us to compute the sample eigenvectors. The algorithm can be used “as is” to compute the federated approximate vertical PCA by projecting the approximate left eigenvector to the data; or as an initialization strategy for federated subspace iteration. For the latter, instead of initializing \({\textbf{G}} ^s_0\) randomly (Algorithm 3, Line 6), \({\textbf{G}} ^s_0\) is computed using the approximate algorithm described here (Algorithm 3, Line 2).

Algorithm 4 describes this approach. The algorithm proceeds as follows: At the clients, a local PCA is computed and the top \(c\cdot k\) eigenvectors are shared with the aggregator, with c a constant multiplicative factor (Line 2). At the aggregator, the local eigenvectors are stacked such that a new approximate covariance matrix \(\hat{{{\textbf{M}}}}\) with \(\dim (\hat{{{\textbf{M}}}})= c \cdot k \cdot S\times m\) is formed. \(\hat{{{\textbf{M}}}}\) is then decomposed using singular value decomposition, leading to a new eigenvector estimate \(\hat{{{\textbf{H}}}}\) (Lines 5–6). At the clients, the feature eigenvector estimate \(\hat{{{\textbf{H}}}}\) can be projected onto the data to form an approximation of the sample eigenvector \({\hat{{\textbf{G}}}}^s\). The vectors \({\hat{{\textbf{G}}}}\) and \({\hat{{\textbf{H}}}}\) represent an “educated guess” of the final singular vectors.
3.4 Federated randomized principal component analysis
Another mitigation strategy for the aforementioned information leakage is the use of randomized SVD. In randomized SVD, a reduced representation of the data is computed and subspace iteration is applied on this reduced data matrix instead of the full data. By using the proxy data, only “reduced” eigenvectors become available at the aggregator, which makes the attack in Sect. 4 impossible, given not too many initial iteration \(I^\prime\) have been executed. Notably, \(I^\prime\) needs to be restricted depending on the number of features in the original data. Algorithm 5 describes how to modify randomized SVD such that it can be run in a federated environment without sharing the random projections of the data or the sample eigenvectors.
We proceed according to Halko et al. (2011) and Galinsky et al. (2016). First, \(I^\prime\) iterations of federated vertical subspace iteration are run using the full data matrices \({\textbf{A}} ^s\). In order to do so, Algorithm 3 is called as a subroutine. The intermediate matrices \({\textbf{H}} _1,\ldots ,{\textbf{H}} _I^\prime\) are stored (Line 1) and concatenated to form \(\bf{P} \in \mathbb {R}^{k\cdot I^\prime \times m}\) (Line 2), and singular value decomposition is used to orthonormalize \(\bf{P}\) (Line 3). The data matrices \({\textbf{A}} ^s\) are then projected onto \(\bf{P}\) to form proxy data matrices \(\hat{{{\textbf{A}}}}^s \in \mathbb {R}^{k\cdot I^\prime \times n}\) (Line 5). Finally, the covariance matrix of the proxy data matrix is computed as \(\hat{{{\textbf{A}}}}^s\hat{{{\textbf{A}}}}^{s\top } \in \mathbb {R}^{k\cdot I^\prime \times k\cdot I^\prime }\) at the clients. The clients send this covariance to the aggregator, which aggregates the covariance matrices \({\textbf{M}} _{{\hat{{\textbf{A}}}}} = \sum _{s} \hat{{{\textbf{A}}}}^s\hat{{{\textbf{A}}}}^{s\top }\) by element-wise addition, computes its eigenvectors \(\textbf{G} _{{\hat{{\textbf{A}}}}}\), and shares them with the clients. The eigenvectors \({\textbf{G}} _{{\hat{{\textbf{A}}}}} \in \mathbb {R}^{k\cdot I^\prime \times k}\) do not reflect properties of the original data, but they can be used to recompute \({\textbf{G}}\) as \({\textbf{G}} = \hat{{{\textbf{A}}}}^{s\top } {\textbf{G}} _{{\hat{{\textbf{A}}}}} \in \mathbb {R}^{n \times k}\). \({\textbf{G}}\) needs to be normalized using the federated orthonormalization subroutine (Algorithm 2). The subroutine returns the correct right singular vectors \({\textbf{G}}\) but only proxy vectors for \({\textbf{H}}\). Therefore, in the last step, \({\textbf{H}}\) can be reconstructed by projecting the data onto \({\textbf{G}}\), aggregating and normalizing \({\textbf{H}} ^s\) at the aggregator, and returning the final left singular vectors \({\textbf{H}}\) to the clients (Lines 18–21).
Algorithm 5 is exactly equivalent to the centralized version, which implies that convergence behaviour and error bounds which have been established for the centralized version (Musco and Musco 2015; Wang et al. 2015) translate to Algorithm 5. The algorithm proceeds in four phases: (1) the initial iterations, (2) the computation and decomposition of the aggregated reduced covariance matrix, (3) the projection and federated orthogonalisation of \({\textbf{G}} ^s\), and (4) the computation and orthonormalization of \({\textbf{H}}\). We have shown that centralized and federated subspace iteration are equivalent (Proposition 2), which extends to the first and the fourth phases of the algorithm. A global covariance matrix can be computed exactly as the sum of the local covariance matrices, which implies that the second phase of the algorithms maintains equivalence. From Proposition 1, it follows that also the third phase maintains equivalence.

3.5 Network transmission costs
The main bottleneck in FL is the amount of data transmitted between the different sites and the number of network communications (Kairouz et al. 2021). The following Proposition 3 specifies these quantities for our federated PCA algorithm. Recall that S, k, m, n, and c denote, respectively, the numbers of sites, eigenvectors, features, samples, and a user-defined constant multiplicative factor.
Proposition 3
Let \({\mathcal {D}}\) be the total amount of data transmitted by the federated SVD algorithm measured in matrix elements, \({\mathcal {N}}\) be the total number of network communications, and I be the total number of iterations of the main while-loop. Let further \(I'\) be the number of initial iteration for randomized SVD and \(k'\) the intermediate dimensionality of the subspace for the approximate algorithm. Then, the following statements hold:
-
If the \({\textbf{G}} _i\) matrices are not orthonormalized, then \({\mathcal {D}}={\mathcal {O}}(I\cdot S\cdot k\cdot m)\) and \({\mathcal {N}}={\mathcal {O}}(I\cdot S)\).
-
If federated Gram–Schmidt orthonormalization is used, then \({\mathcal {D}}={\mathcal {O}}(I\cdot (S\cdot k\cdot m + k^2))\) and \({\mathcal {N}}={\mathcal {O}}(I\cdot S\cdot k)\).
-
If federated randomized subspace iteration is used, then \({\mathcal {O}}((S\cdot (I'+1) \cdot k \cdot m)+(k \cdot I')^2)\) and \({\mathcal {N}}={\mathcal {O}}((I'+ 2)\cdot S)\).
-
Approximate initialization itself has a complexity of \({\mathcal {D}}={\mathcal {O}}(S\cdot k \cdot c \cdot m)\) and \({\mathcal {N}}={\mathcal {O}}(S)\), hence the other algorithms remain in the same complexity class if used in combination with approximate initialization.
Proof
In each iteration i of our federated vertical subspace iteration algorithm, the matrices \({\textbf{H}} _i^s\in \mathbb {R}^{m\times k}\) have to be sent from the clients to the aggregator and the matrix \({\textbf{H}} _i\in \mathbb {R}^{m\times k}\) has to be sent back to the clients. In iteration i, the amount of transmitted data and the number of communications due to \({\textbf{H}} _i\) is hence \({\mathcal {O}}(S\cdot k\cdot m)\) and \({\mathcal {O}}(S)\), respectively. For orthonormalizing the eigenvector matrices \({\textbf{G}} _i\in \mathbb {R}^{n\times k}\), we need to transmit a data volume of \({\mathcal {O}}(S\cdot k^2)\) and the number of communications increases to \({\mathcal {O}}(S\cdot k)\). By summing over the iterations i, this yields the statement of the proposition. In the randomized iteration the first \(I'\) iterations have the same communication complexity that regular subspace iteration. Then the dimensionality of the matrix is reduced to \(k\cdot I'\times n\) and the decomposition of \({\textbf{M}} _{{\hat{{\textbf{A}}}}}\) has transmission complexity \((k\cdot I')^2\). An additional communication of the final \({\textbf{H}}\) is required which costs \(S \cdot k \cdot m\). Thereby, the total complexity is \({\mathcal {D}}= {\mathcal {O}}(I'\cdot S \cdot k \cdot m + (I'\cdot k)^2 + S \cdot k \cdot m) = {\mathcal {O}}((S\cdot (I'+1) \cdot k \cdot m)+(k \cdot I')^2)\). Approximate initialization has a complexity of one round of subspace iteration, as \({\textbf{H}} _i\) needs to be communicated once to the aggregator and back. The complexity classes hence remain the same. \(\square\)
If our algorithms are used, the overall volume of transmitted data is hence independent of the number of samples n and can be executed in a constant number of communication rounds. This is especially important in the intended GWAS setting, since the number of samples and features may be very large (Li et al. 2016; Londin et al. 2010). Moreover, k is small (typically, \(k=10\) is used for GWAS PCA), which implies that the additional factor k in the complexities of \({\mathcal {D}}\) and \({\mathcal {N}}\) can be neglected. Therefore, using the suggested scheme is preferable over sending the eigenvectors to the aggregator for orthonormalization, both in terms of privacy and expected transmission cost. (In practice, it is advisable to perform the orthonormalization only at the end.) The algorithm by Guo et al. (2012) has a complexity of \({\mathcal {D}}={\mathcal {O}}(I\cdot S\cdot k)\) and \({\mathcal {N}}= {\mathcal {O}}(I\cdot S)\) per eigenvector. The algorithm by Chai et al. (2021) has a data transfer complexity of \({\mathcal {D}}={\mathcal {O}}(n \cdot m)\), with \({\mathcal {N}}= {\mathcal {O}}(S)\). The use of randomized SVD additionally partially removes the dependency of the algorithm on the number of SNPs/features, which can be quite large in practice. (The worst-case complexity does not change due to the first iterations.) Additionally, only a few iterations of the true feature eigenvectors are transmitted. Therefore, randomized SVD is preferable in terms of transmission cost.
3.6 Summary and applications
To conclude this section, we provide a brief summary of the main points and introduce a naming scheme for the configurations evaluated in Sect. 5. We presented federated vertical subspace iteration with random (RI-FULL) initialization. To avoid the sharing of the sample eigenvector matrix, we introduced federated Gram–Schmidt orthonormalization (FED-GS) which can be run at every iteration, but should be run only at the end. In order to speed up the computation in terms of communication rounds, we suggest to use a modified version of the approximate algorithm (AI-ONLY) by Balcan et al. (2014) as an initialization strategy for federated subspace iteration (AI-FULL). To reduce the transmitted data volume and the sharing of the feature eigenvectors, we suggest to use federated randomized subspace iteration (RANDOMIZED). GUO is the reference algorithm. We summarize the asymptotic communication costs in Table 2. We show that federated and centralized subspace iteration are equivalent and explain how this property propagates to the randomized algorithm.
The results returned by the algorithms are the left singular vectors (which are identical for all participants and known to all participants), the singular values (also known to all participants), and the partial right singular vectors, where each part is known only to its owner. The singular vectors can be used to compute the corresponding reduced representation of the data locally, by projecting the data onto the leading eigenvectors, sometimes called the principal components. These projections can technically be shared with all other participants if desired. Thereby, at least in principle, all standard functionalities are covered by the presented algorithms. The next section will cover some privacy aspects related to these results.
4 Privacy
In this section, we describe how the iterative process discloses the covariance matrix when using sufficiently many iterations (Sect. 4.1). Next, we illustrate the problem using a simulation study (Sect. 4.2) and then discuss how it can be mitigated with the algorithms introduced above and via SMPC and DP (Sect. 4.3).
4.1 Iterative leakage of the covariance matrix
Iterative leakage at the aggregator might disclose the entire covariance matrix during the execution of the algorithm, as many updates of the variables become available. Figure 3 visualizes the update process in power iteration, and the information used to reconstruct a single row of the covariance matrix at one iteration. Notably, the aggregated vector \({\textbf{H}} _i\) becomes known in clear text at the aggregator in every iteration. The aggregator can store the sequence of vectors \({\textbf{H}} _i\). In the following, we will show how it is possible to construct a system of linear equations which will leak the covariance matrix. For the sake of this description, we will assume the eigenvector \({\textbf{H}} _i\) is updated as \({\textbf{H}} _i= {\textbf{K}} {\textbf{H}} _{i-1}\), where \({\textbf{K}} ={\textbf{D}} ^\top {\textbf{D}}\) is the feature-by-feature covariance matrix of the federated data matrix \({\textbf{D}}\), which are both unknown to the aggregator. This is equivalent to the two-step update from the aggregator’s perspective, but improves the readability.
Proposition 4
Let \({\textbf{D}} \in \mathbb {R}^{n\times m}\) be the data matrix and denote \({\textbf{K}} ={\textbf{D}} ^\top {\textbf{D}}\) the feature-by-feature covariance matrix, which is unknown to the aggregator. Let k be the number of eigenvectors retrieved. When applying federated subspace iteration, the aggregator can reconstruct \({\textbf{K}}\) after m/k distinct eigenvector updates by solving a system of linear equations of the form \({\textbf{K}} _{l,\bullet }{\textbf{A}}={\textbf{b}}\) for each row \({\textbf{K}} _{l,\bullet }\) of \({\textbf{K}}\), where \({\textbf{A}}\in \mathbb {R}^{m\times m}\) and \({\textbf{b}}\in \mathbb {R}^{m}\) are known parameters.
Proof
Let \({\textbf{K}} _{l,\bullet }\) denote a row of the covariance matrix \({\textbf{K}} \in \mathbb {R}^{m\times m}\). First, we show how series \(({\textbf{H}} ^i_{\bullet , 1})_{i=1}^m\) of m updates of the first eigenvector can be used to retrieve the row \({\textbf{K}} _{l,\bullet }\) of \({\textbf{K}}\). Subsequently, we show that m/k updates are sufficient if all k eigenvectors are used.
Since \({\textbf{K}} _{l,\bullet }\) is a row vector of length m, one needs m equations, which can be derived from m consecutive updates of the column vector \({\textbf{H}} ^i_{\bullet , 1}\). The aggregator can store the consecutive updates of \({\textbf{H}} ^i_{\bullet , 1}\) and, for each i, store an equation of the form \({\textbf{K}} _{l,\bullet } {\textbf{H}} ^{i-1}_{\bullet , 1}=H^i_{l,1}\). After m iterations, the aggregator is able to formulate the following fully determined system of linear equations, given that the eigenvectors have not converged:
In order to reduce the number of required iterations, the aggregator can use all vectors in \({\textbf{H}}\) to formulate the linear system and thereby divide the number of required iterations by k:
The rows of \({\textbf{K}}\) can be computed simultaneously, by forming a system for all \({\textbf{K}} _{l, \bullet }\) at the same time. Therefore, in theory, this means that after m/k iterations, one has the full system and can solve it as
which completes the proof of the proposition. \(\square\)
These theoretical results require to invert \({\textbf{A}}\), which may pose a problem in numerical applications, especially once the \({\textbf{H}} ^i\) grow large. By using a linear least squares solver, the inversion of the matrix \({\textbf{A}}\) can be prevented at the cost of possibly sub-optimal solutions. Furthermore, in practice, more care needs to be taken when constructing the system, because, once converged, the eigenvectors do not provide a new equation to be added to the system anymore and hence lead to a singular system.
4.2 Covariance reconstruction experiment
We implemented the covariance reconstruction scheme and applied it on small example data, demonstrating its practicality. Using the breast cancer dataset and the diabetes data set from the UCI repository (Dua and Graff 2017), which have 442 and 569 samples and 10 and 30 features, respectively, we computed the centralized covariance matrix which we aim to reconstruct. Then, we ran federated subspace iteration and recorded the eigenvector updates. After m/k iterations, we used the recorded matrices to form the linear system described in Sect. 4.1. Instead of inverting the matrix as described in Eq. 6, we used a linear least squares solver (scipy.lstsq) to compute the solution. We then computed the Pearson correlation between the true and the reconstructed covariance matrix, with outcomes of 1.0 and 0.997, (1.0 indicates perfect reconstruction) in negligible time (see Table 3). The results imply that the attack described above constitutes a practically relevant privacy risk and hence needs to be mitigated.
4.3 Mitigation strategies and remaining privacy concerns
Here, we briefly discuss the remaining privacy concerns with our algorithm. To do so, we distinguish two types of disclosure: information intentionally published as a result of the algorithm and information that is somehow obtained by any party (including outsiders) in the course of the execution, but is not part of the results. In this article, we assume honest-but-curious participants. This means that the participants execute the algorithm correctly but try to infer as much information as possible about the other participants. All participants want to obtain the correct and complete result, which we define as the complete top k feature eigenvectors, the singular values, and the top k partial sample eigenvectors. An unintended information leak is hence information that can be inferred beyond the top k singular vectors. Such a leak would for instance be the disclosure of the exact covariance matrix, because it would allow the computation of additional singular vectors. We begin this part with a brief presentation of privacy-enhancing techniques.
4.3.1 Secure multiparty computation
SMPC allows multiple parties to compute a function collaboratively without sharing their secret inputs. An established concept is secret sharing, where the input values are split into shares, which disclose nothing about the original value. Modular arithmetic is used to mask the contributions. The N participants determine a fixed prime number p. Then, they split their secret \(s_n \in \mathbb {Z_p}\) by uniformly generating random numbers \(r_{n,i}\), for \(i \in [N-1]\). The last random share is computed as \(r_{n,N} = s_n-\sum _{i=1}^{N-1} r_{n,i} \mod p\). \(N-1\) of their shares \(r_{n,i}\) are sent to the \(N-1\) players and the Nth share remains with them. Each player computes the sum over the shares, including their own, as \(r_i=\sum _{n=1}^{N} r_{n,i}\) and redistributes \(r_i\) to all other players. Then, every participant can compute the output \(o=\sum _{i}^{N} r_i = \sum _{n=1}^{N} s_n\) as the sum over all \(s_n\), while the private inputs are kept safe. While this protocol is correct and secure, it is not tolerant to the event that one of the participants leaves during the execution of the protocol (Cramer et al. 2015). If this is a concern, more fault-tolerant methods such as Shamir’s secret sharing algorithm can be used (Cramer et al. 2015). In this protocol, the participants choose a secret polynomial f of degree k. This polynomial evaluates to the secret value at f(0). Instead of splitting the data, the participants distribute points of the polynomial. The secret can be reconstructed using \(k+1\) points of the polynomial with the help of Lagrangian multipliers. In this protocol, \(p>k\) players need to remain in the protocol, but k may be smaller than the number of participants (\(k<n\)). Float values can be processed using fixed-point arithmetic (Ryffel et al. 2018).
4.3.2 Differential privacy
DP is a rigorous statistical framework designed to statistically query datasets without disclosing too much about their contents (Dwork and Roth 2013). It hinges on the design of a mechanism \({\mathcal {M}}\) which yields very similar results when applied to two adjacent datasets. DP can be achieved through different techniques, e.g., the addition of noise to the input, to the parameters, or to the output of the algorithm. The noise is scaled to the sensitivity of the mechanism \({\mathcal {M}}\), which is defined as the maximal difference of the outputs when the algorithm is run on two adjacent datasets. Since DP exhibits “closure under post-processing” (Dwork and Roth 2013), any computation performed on the result of a differentially private analysis remains differentially private. DP is hence a framework that allows the publication of results which themselves constitute potential privacy leakages (such as the right or left singular vectors in the case of federated SVD).
4.3.3 Properties of our algorithms limiting information leakage
The design of our algorithm includes some sensible defaults that limit information leakage. If complete information were available (including all singular vectors and values), lossless reconstruction of the data matrix would be possible. Consequently, for the privacy-preserving publication of the results, information has to be omitted or masked. In Sect. 3, we developed an algorithm which returns the top k singular subspace. This is often called a truncated SVD. It prevents the lossless reconstruction of the data but may result in a very good approximation, depending on the data and the number of eigenvectors k. Additionally, we discussed in Sect. 1 that the user can and should at least omit the sample-associated singular vectors. Therefore, the most straightforward measures to avoid excessive data leakage have already been included in the design of the algorithms.
We have established that the full global covariance matrix can be reconstructed after sufficiently many iterations. Once the covariance matrix has been reconstructed, one can compute all singular vectors, i.e., , more than the top k. The randomized Algorithm 5 shares the initial eigenvector updates, but then shares only proxy eigenvectors, whose entries do not correspond to real features in the data, effectively reducing the number of useful iterations for the aggregator to a constant number \(I^\prime\). The first iterations do allow to estimate a proxy covariance matrix, but so do the top k singular vectors of the result, to higher accuracy. The limitation of the number of iterations hence improves the privacy of the randomized algorithm compared to classical federated power iteration. In the envisaged use case, we assume \(I'\cdot k \ll \min (d,n)\), preventing the attack in practice.
4.3.4 Remaining privacy concerns: intermediate parameters
Even with the limited number of information exchanges, certain intermediate parameters are disclosed to all participants in order to compute the correct result—in particular, the feature eigenvectors updates and the reduced representation of the data. This also poses a certain risk to the participants, which can be mitigated using SMPC and DP.
SMPC
Disclosure of local intermediates in the presented algorithms can be prevented by using additive SMPC, which ensures that all additive aggregation steps in the algorithm can be performed securely. In the algorithms presented in this article, almost all aggregation steps use the sum operation as the first step in the aggregation routines (Lines 6, 15 and 22 in Algorithm 2,ine 16 in Algorithm 3, Line 5 in Algorithm 5). The only exception is Line 5 in Algorithm 4 (vertical stacking of partial feature eigenvectors). Avoiding the use of vertical stacking here would require to expand the subspaces to approximate covariance matrices \({\textbf{V}} ^s{\textbf{V}} ^{s\top }\), which is problematic in the high-dimensional case due to the space requirements of these matrices. The naïve communication would increase from \({\mathcal {O}}(S)\) to \({\mathcal {O}}(S^2)\), but with smarter schemes (e.g., Shamir’s scheme (Cramer et al. 2015)), the overhead can be limited.
Unfortunately, in the case of power iteration or low dimensional data, the securely aggregated intermediate parameters can still be attacked using the scheme described above, as the aggregated updates (i.e., , the 10 initial updates of \({\textbf{H}}\), and the intermediate covariance matrix \({\textbf{M}}\)) still become available in clear text at the aggregator. However, the individual clients have masked their individual local updates (and hence their local covariance matrices). The SMPC schemes we presented assume a peer-to-peer communication between the clients. This means, it is possible to complete the protocol without the involvement of a third party aggregator and therefore nothing is leaked to outside parties.
DP
The algorithm can be divided into different phases for which DP algorithms have been developed. Hardt and Price (2013) have determined a noise scheme for the power method, whereas Chaudhuri et al. (2013) provide the noise variance for the covariance perturbation. Consequently, both phases of the algorithm can be transformed into differentially private mechanisms. By the composition theorem (Dwork and Roth 2013), the result will also be a DP mechanism. Alternatively, if the parties have no concerns over sharing the unmasked intermediate parameters, the output can be masked using a noise scheme such as the one proposed by Asi and Duchi (2020). This would provide DP for outside third parties, but would not protect the participants from internal attacks. The noise scheme suggested by Grammenos et al. (2020) cannot be applied directly, as the algorithm is designed for a single pass over the data.
Hybrid methods
SMPC and DP can also be applied in combination, for instance to yield a certain \(\epsilon\) globally, instead of enforcing a local \(\epsilon\). Small noise in combination with the limited extra disclosure due to the low number of iterations might provide a good trade-off between privacy and utility.
4.3.5 Remaining privacy concerns: publication of the projections
Lastly, a popular application of SVD is PCA, which includes the projection of the data onto the leading eigenvectors to obtain a low-dimensional representation of the data. Although this is a publication of more information than we defined as the output of the algorithm at the beginning of this section, we deem it important to comment on this, due to the popularity of the procedure. In the federated use case, the projection can be performed entirely at the client for both singular subspaces. The clients can then choose to share the projections. For some applications—e.g., if the reduced representation is used as input to a neural network—sharing may not be necessary. For other applications such as the visual control of batch effects, users may choose to send the projections. In this case, users should keep in mind that computing projections using the perturbed eigenvectors does not fall under the closure under postprocessing because the data is reused. If a client is in possession of both the eigenvector and the low-dimensional embedding, they can be used to reconstruct an approximation of the data (Hartebrodt and Röttger 2022). Even if only the projections and no global eigenvectors were known to the participants (a hypothetical scenario with the presented algorithm), users could use the inverted data matrix to reconstruct the eigenvectors based on their own data. Therefore, the publication of the projections presents a risk which is hard to mitigate even with DP.
4.4 Concluding remarks on privacy concerns
To conclude, the proposed randomized scheme does not achieve perfect privacy in the sense of an oracle which only outputs the required result, as the aggregated initial \(I'\) iterations and the reduced covariance matrices are known to the participants. However, it reasonably reduces the available information beyond the final results and can be easily combined with DP and SMPC. Integrating DP and SMPC in the algorithms presented in this paper is left for future work.
5 Empirical evaluation
5.1 Test datasets
To evaluate our federated PCA algorithm, we used several publicly available datasets: chromosome 1 and 2 from a genetic dataset from the 1000 Genomes Project (The 1000 Genomes Consortium, Auton 2015), the MNIST dataset of handwritten digits (LeCun et al. 2005), the MOVIELENS 25 M dataset (Harper and Konstan 2015), and a high-throughput sequencing dataset (Benz et al. 2022) containing single-cells transcriptomics (MULTIOME-RNA) data. Table 4 provides an overview of the test datasets.
The two genetic datasets contain minor allele frequencies for 2502 individuals (samples). After applying standard pre-processing steps (MAF filtering, LD pruning), we created 3 dataset versions for each chromosome, with 0.1 million, 0.5 million, and \(>1\) million SNPs, respectively. To the best of our knowledge, publicly available genetic data sets with large numbers of patients are not readily available. MNIST contains 60000 grayscale images of handwritten numerals (samples), each of which has 784 pixels (features). MOVIELENS contains user ratings for roughly 60000 movies (features) from about 162000 users (samples). We used two versions of this dataset: the original sparse dataset and a scaled dense version with unit variance and zero mean. MULTIOME-RNA containes gene expression counts for 23418 genes (features) and 105942 cells. The experiments on MNIST, MOVIELENS, and MULTIOME demonstrate thei usefulness of the algorithms for applications beyond federated GWAS.
5.2 Compared methods
We compare several configurations of our algorithms: Federated subspace iteration with random initialization (RI-FULL), federated subspace iteration with approximate initialization (AI-FULL), and federated randomized subspace iteration (RANDOMIZED). Federated subspace iteration with federated orthonormalization in every round (FED-GS) is extremely communication-inefficient; the \(c\cdot k\) additional rounds per iteration have proven to be a major bottleneck in preliminary experiments. Therefore, we omit this algorithm from the empirical evaluation. We compare our algorithms to a centralized baseline and to the algorithm presented by Guo et al. (2012), denoted GUO, as they present a solution which omits the covariance matrix and a way to deal with vertical data partitioning. However, GUO shares the right singular vectors \({\textbf{G}}\) and all updates of the left singular vectors \({\textbf{H}}\) with the aggregator. This should be avoided in federated GWAS, as emphasized in Sects. 3 and 4. We tested other configurations, including the use of approximate initialization for randomized PCA, but excluded them in this article as they did not bring a gain in performance in practice.
For all compared methods, we set the convergence criterion in Eq. 5 to \(t=10^{-9}\), which corresponds to a change of the angle between two consecutive eigenvectors updates of about 0.0026 degrees. Note that this angle does not equal the angle w.r.t. centrally computed eigenvectors, which we used as a test metric for measuring the quality of the compared methods (see next subsection). To avoid excessive computations, we used a restricted experimental setup for the tests on the MOVIELENS and MULTIOME datasets: On these larger datasets, we only compared RANDOMIZED (which showed the overall best performance among all tested federated algorithms on the other datasets) against the centralized baseline. For MOVILENS, we also compared the runtime of RANDOMIZED to the reported runtime of the approach by Chai et al. (2021) on the same dataset.
5.3 Test metrics
For measuring the quality of the compared methods, we computed the angles between the eigenvectors obtained from a reference implementation of a centralized PCA and their counterparts computed in a federated fashion. An angle of 0 between two eigenvectors of the same rank is the desired result. As a reference, we chose the version implemented in scipy.sparse.linalg, which internally interfaces LAPACK. The amount of transmitted data is estimated by calculating the number of transmitted floats and multiplying it by a factor of 4 bytes (single precision IEEE 754). We choose this metric to remain agnostic with respect to the transmission protocol. Time measurements are wall clock times using Python’s time module. We chose to measure the runtime for matrix operations only, as they are the most important contributors to the overall runtime apart from communication-related runtime.
5.4 Implementation, availability, and hardware specifications
All methods are implemented in Python, using mainly, but not exclusively numpy and scipy. They are available online at https://github.com/AnneHartebrodt/federated-svd. The simulation tests were run on a compute server with 48 virtual CPUs (2.5 GHz) and 502 GB available RAM. Tests for Table 6 were run on a CPU server with 96 virtual CPUs (2.8GHz) and 500GB RAM, restricted to use 22 CPUs on the same socket to reduce data copying and waiting times, unless otherwise specified. A federated tool compatible with the FeatureCloud ecosystem (Matschinske et al. 2023) is available at https://featurecloud.ai/app/federated-svd. The corresponding source code can be found at https://github.com/AnneHartebrodt/fc-federated-svd. We also created an AIMe report (Matschinske et al. 2021) to promote accessibility of machine learning research (Hartbrodt 2022).
5.5 Convergence behavior
To test the convergence behavior of the compared federated algorithms, we split the genetic datasets into 5 equally sized chunks and the MNIST dataset into 5 and 10 chunks. For every algorithm, we then recorded the angles between the first 10 eigenvectors and the fully converged references at each iteration averaged across 10 runs.
Figures 4 and 5 show the convergence behaviours of the tested algorithms on MNIST and the genetic datasets. Each panel contains a line plot with one line per tested algorithm. On the x-axis, we show the number of iterations; on the y-axis, the angle of the eigenvector w.r.t. the reference in function of the number of iterations. Each column of panels is dedicated to one eigenvector. In Fig. 4, the rows contain the results for a test with different numbers of sites. In Fig. 5 the rows are split between a varying number of features (SNPs).
Convergence behaviour of federated algorithms on the MNIST data. The number of iterations is shown on the x-axis, the angle between the federated eigenvectors and the centralized eigenvector is shown on the y-axis. The omitted eigenvectors show similar behaviors. The top and bottom rows show the convergence for 5 and 10 simulated sites, respectively
Convergence behaviour of federated algorithms on Chromosome 1 and 2 of the 1000 Genomes data. The number of iterations is shown on the x-axis, the angle between the federated eigenvector and the centralized eigenvector is shown on the y-axis. The omitted eigenvectors show similar behaviors. The three rows in each plot show the results for 0.1 million, 0.5 million, and all SNPs in the datasets
The most important result is that, for all algorithms, the eigenvectors perfectly converge to the reference eventually. For small k, AI-FULL starts with a good approximation of the eigenvectors and therefore converges very quickly, but for higher ranks, the initial approximation is on average not better than a random guess (see Fig. 5, Eigenvector 10). GUO shows good convergence behavior once the iteration has started. However, unlike our algorithms RI-FULL, AI-FULL, and RANDOMIZED, GUO computes the eigenvectors sequentially. This means that eigenvector k has to converge before the computation of the eigenvector \(k+1\) starts, because of which all plots except the one for the first eigenvector start with a horizontal line for GUO. RANDOMIZED initially shows the same convergence behavior as RI-FULL, but immediately jumps to the final approximation at iteration \(I'+1=11\) and hence consistently exhibits the same fast convergence. For low-ranking eigenvectors, the approximate initialization speeds-up the computations, because these eigenvectors can be well approximated and the computations hence already start with very small deviations from the reference. The gain decreases in higher dimensions. In sum, RANDOMIZED shows the overall best convergence behavior across all datasets and dimensions.
The number of sites does not influence the convergence behavior, as can be seen in the tests on the MNIST data, where the convergence curves (Fig. 4) and the required number of iterations (Fig. 7) are similar for the simulations with 5 clients and 10 clients. Varying the number of SNPs (features) in the genetic datsets does not show a clear effect. Although, in the convergence plots (Fig. 5), the algorithms seem to converge faster on the larger datasets, the overall number of iterations in Fig. 6 does not confirm this trend. The reason for this is the dependence of the convergence speed on the eigengaps (the difference between two consecutive eigenvalues), which are inherent properties of each dataset: The smaller the eigengaps, the worse the convergence behavior (Li et al. 2021). Table 5 shows the eigengaps for Chromosome 2. The higher-ranking eigengaps are generally quite small, indicating generally bad convergence for all datasets. Eigengap 8 for the dataset containing 1140556 SNPs is particularly small, which could explain the especially poor convergence.
5.6 Scalability
To gauge the scalability of the methods with respect to runtime and transmission cost, we recorded the number of iterations until convergence, the runtime for matrix operations, and the estimated total amount of transmitted data for the selected algorithms. The results of the direct comparison of all algorithms for the genetic datasets and MNIST are summarized in Figs. 6 and 7. There are three plots per dataset, which contain the number of iterations until convergence, the overall runtimes for matrix operations, and the estimated transmitted data. The tested algorithms are shown on the x-axes. In Fig. 6, for each algorithm and chromosome, there are three boxplots, one for each test with different numbers of SNPs. In Fig. 7, there are two boxplots for each algorithm, corresponding to splits of the data into 5 and 10 sites.
Iterations, runtimes for matrix computations, and total transmitted data for the two genetic datasets. On the x-axis, we show the tested algorithms, on the y-axis, the measured performance indicators. Single horizontal bars indicate that there is no spread in the data. For each algorithm and dataset, there are three boxplots, representing the number of SNPs in the data
Iterations, runtime for matrix computations, and total transmitted data for the MNIST dataset. On the x-axis, we show the tested algorithms, on the y-axis, the measured performance indicators. Single horizontal bars indicate that there is no spread in the data. For each algorithm and dataset, there are two boxplots, representing the number of simulated clients
The amount of transmitted data is the smallest for RANDOMIZED, followed by GUO, AI-FULL, and RI-FULL (the latter two with a significant distance). RANDOMIZED also spends the least time on the matrix operations, which are the major contributors to the runtime. The number of required iterations is the smallest for RANDOMIZED, followed with large gap by AI-FULL, RI-FULL, and GUO on the last place. Since the required number of iterations correspond to the number of communication steps, this factor significantly contributes to the overall runtime. Notably, the number of iterations for RANDOMIZED is constant on all datasets, indicated by the horizontal bars in the plots. Overall, RANDOMIZED performs the best in all three measured categories. Generally, the bottleneck in FL is the number of transmission steps during the learning process, as this involves network communication. However, with increasing dataset size, also the reduction in local runtime shows considerable impact on the overall runtime.
5.7 Additional tests on MOVIELENS and MULTIOME datasets
Table 6 shows additional runtime and convergence results for RANDOMIZED on the MOVIELENS and the MULTIOME datasets. With respect to the convergence, the most important result is that even higher-order eigenvectors can be retrieved to high accuracy. Our algorithm requires on average 18 min to compute \(k=256\) eigenvectors for the MOVIELENS dataset using scaled and centered data and 9 min using the original sparse data. According to their own benchmark, the algorithm by Chai et al. (2021) requires 222 min to compute the same number of eigenvectors for MOVIELENS. Since Chai et al. (2021) simulated their PCA on a computer with 8 CPUs (3.6 GHz), we carried out additional tests with the number of CPUs (2.8 GHz) restricted to 8. Even with this setup, RANDOMIZED still achieves a runtime of \(25\ll 222\) min on average. Therefore, our algorithm achieves a significant speed-up.
6 Conclusions
In this paper, we presented an improved federated SVD algorithm, which is applicable to both vertically and horizontally partitioned data and, at the same time, increases the privacy compared to previous solutions. The algorithm is applicable on sparse and dense matrices. Although our algorithm is motivated by the requirements of population stratification in federated GWAS, it is generically applicable. We proved that a first version of our algorithm is equivalent to a state-of-the-art centralized SVD algorithm and demonstrated empirically that it indeed converges to the centrally computed solutions. Subsequently, we improved the algorithm by including techniques from other federated and centralized algorithms to increase scalability and reduce the number of required communications.
There are two key advantages of our algorithm: Firstly, unlike in existing federated PCA algorithms, the sample eigenvectors remain at the local sites, due to the use of fully federated Gram–Schmidt orthonormalization. This significantly improves the privacy of the algorithm. Secondly, the algorithm limits the amount of transmitted data via smart initialization and data approximation. This leads to a better scalability and further prevents information leakage. In particular, the transmission cost is no longer dependent on the number of samples and only partially depends on the number of features.
References
Al-Rubaie M, Wu PY, Chang JM, et al (2017) Privacy-preserving PCA on horizontally-partitioned data. In: 2017 IEEE conference on dependable and secure computing, pp 280–287. https://doi.org/10.1109/DESEC.2017.8073817
Asi H, Duchi JC (2020) Instance-optimality in differential privacy via approximate inverse sensitivity mechanisms. In: Larochelle H, Ranzato M, Hadsell R, et al (eds) Advances in neural information processing systems, vol 33. Curran Associates, Inc., pp 14106–14117. https://proceedings.neurips.cc/paper/2020/file/a267f936e54d7c10a2bb70dbe6ad7a89-Paper.pdf
Balcan MF, Kanchanapally V, Liang Y et al (2014) Improved distributed principal component analysis. In: Advances in neural information processing systems 4(January), pp 3113–3121. arXiv: org/abs/1408.5823
Balcan MF, Du SS, Wang Y et al (2016) An improved gap-dependency analysis of the noisy power method. J Mach Learn Res 49(June):284–309 (http://arxiv.org/1602.07046)
Benz A, Chow A, Burkhardt D et al (2022) Open problems: multimodal single-cell integration. https://kaggle.com/competitions/open-problems-multimodal
Chai D, Wang L, Zhang J et al (2021) Practical lossless federated singular vector decomposition over billion-scale data. arXiv: org/abs/2105.08925
Chaudhuri K, Sarwate AD, Sinha K (2013) A near-optimal algorithm for differentially-private principal components. J Mach Learn Res 14:2905–2943
Chen X, Lee JD, Li H et al (2021) Distributed estimation for principal component analysis: a gap-free Approach. arXiv: org/abs/2004.02336
Cho H, Wu DJ, Berger B (2018) Secure genome-wide association analysis using multiparty computation. Nat Biotechnol 36(6):547–551. https://doi.org/10.1038/nbt.4108
Cramer R, Damgård IB et al (2015) Secure multiparty computation. Cambridge University Press
Dua D, Graff C (2017) UCI machine learning repository. http://archive.ics.uci.edu/ml
Dwork C, Roth A (2013) The algorithmic foundations of differential privacy. Found Trends Theor Comput Sci 9(3–4):211–407. https://doi.org/10.1561/0400000042
Froelicher D, Cho H, Edupalli M et al (2023) Scalable and privacy-preserving federated principal component analysis. arXiv: org/abs/2304.00129
Galinsky KJ, Bhatia G, Loh PR et al (2016) Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. Am J Hum Genet 98(3):456–472. https://doi.org/10.1016/j.ajhg.2015.12.022
Gauch HG, Qian S, Piepho HP et al (2019) Consequences of PCA graphs, SNP codings, and PCA variants for elucidating population structure. PLoS ONE 14(6):1–26. https://doi.org/10.1371/journal.pone.0218306
Gonen A, Murphy K, Schölkopf B (2018) Smooth sensitivity based approach for differentially private principal component analysis. J Mach Learn Res 1:1–48
Grammenos A, Smith RM, Crowcroft J et al (2020) Federated principal component analysis. In: Larochelle H, Ranzato M, Hadsell R et al (eds) Advances in neural information processing systems, vol 33. Curran Associates, Inc., pp 6453–6464. https://proceedings.neurips.cc/paper/2020/file/47a658229eb2368a99f1d032c8848542-Paper.pdf
Guo YF, Lin X, Teng Z et al (2012) A covariance-free iterative algorithm for distributed principal component analysis on vertically partitioned data. Pattern Recognit 45(3):1211–1219. https://doi.org/10.1016/j.patcog.2011.09.002
Hadri B, Ltaief H, Agullo E et al (2010) Tile QR factorization with parallel panel processing for multicore architectures. In: 2010 IEEE international symposium on parallel distributed processing (IPDPS), pp 1–10. https://doi.org/10.1109/IPDPS.2010.5470443
Halko N, Martinsson PG, Shkolnisky Y et al (2010) An algorithm for the principal component analysis of large data sets. arXiv: org/abs/1007.5510
Halko N, Martinsson PG, Tropp JA (2011) Finding structure with randomness: probabilistic algorithms for constructing approximate matrix decompositions. SIAM Rev 53(2):217–288. https://doi.org/10.1137/090771806
Hardt M, Price E (2013) The noisy power method: a meta algorithm with applications. arXiv arXiv: org/abs/1311.2495
Harper FM, Konstan JA (2015) The MovieLens datasets: history and context. ACM Trans Interact Intell Syst. https://doi.org/10.1145/2827872
Hartbrodt A (2022) Federated singular value decomposition for high dimensional data [AIMe lP0kqT]. https://aime.report/lP0kqT
Hartebrodt A, Röttger R (2022) Federated horizontally partitioned principal component analysis for biomedical applications. Bioinform Adv. https://doi.org/10.1093/bioadv/vbac026
Hartebrodt A, Nasirigerdeh R, Blumenthal DB et al (2021) Federated principal component analysis for genome-wide association studies. In: IEEE international conference on data mining (ICDM) 2021. https://doi.org/10.1109/ICDM51629.2021.00127
Hoemmen M (2011) A communication-avoiding, hybrid-parallel, rank-revealing orthogonalization method. In: 2011 IEEE international parallel distributed processing symposium, pp 966–977. https://doi.org/10.1109/IPDPS.2011.93
Imtiaz H, Sarwate AD (2018) Differentially private distributed principal component analysis. In: 2018 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 2206–2210. https://doi.org/10.1109/ICASSP.2018.8462519
Imtiaz H, Mohammadi J, Sarwate AD (2019) Distributed differentially private computation of functions with correlated noise, pp 1–40. arXiv: org/abs/1904.10059
Joliffe IT (2002) Principal component analysis. Springer. https://doi.org/10.1007/b98835
Kairouz P, McMahan HB, Avent B et al (2021) Advances and open problems in federated learning. Found Trends Mach Learn 14(1–2):1–210. https://doi.org/10.1561/2200000083
Kargupta H, Huang W, Sivakumar K et al (2001) Distributed clustering using collective principal component analysis. Knowl Inf Syst. https://doi.org/10.4324/9781315799476-12
LeCun Y, Cortes C, Burges CJ (2005) MNNIST database of handwritten digits. http://yann.lecun.com/exdb/mnist/
Lei Q, Zhong K, Dhillon IS (2016) Coordinate-wise Power Method. In: Lee D, Sugiyama M, Luxburg U, et al (eds) Advances in neural information processing systems, vol 29. Curran Associates, Inc., pp 2064–2072, https://proceedings.neurips.cc/paper/2016/file/8b4066554730ddfaa0266346bdc1b202-Paper.pdf
Li X, Wang S, Chen K et al (2021) Communication-efficient distributed SVD via local power iterations. arXiv: org/abs/2002.08014, [cs, math, stat]
Li Y, Byun J, Cai G et al (2016) FastPop: a rapid principal component derived method to infer intercontinental ancestry using genetic data. BMC Bioinform 17(1):1–8. https://doi.org/10.1186/s12859-016-0965-1
Liu B, Tang Q (2019) Privacy-preserving decentralised singular value decomposition. https://eprint.iacr.org/2019/1346
Liu Y, Chen C, Zheng L, et al (2020) Privacy preserving PCA for multiparty modeling. arXiv: org/abs/2002.02091
Londin ER, Keller MA, Maista C et al (2010) CoAIMs: a cost-effective panel of ancestry informative markers for determining continental origins. PLoS ONE 5:e13443. https://doi.org/10.1371/journal.pone.0013443
Matschinske J, Alcaraz N, Benis A et al (2021) The AIMe registry for artificial intelligence in biomedical research. Nat Methods. https://doi.org/10.1038/s41592-021-01241-0
Matschinske JO, Späth J, Bakhtiari M et al (2023) The FeatureCloud platform for federated learning in biomedicine: unified approach. J Med Internet Res 25:e42621
Mothukuri V, Parizi RM, Pouriyeh S et al (2021) A survey on security and privacy of federated learning. Future Gener Comput Syst 115:619–640. https://doi.org/10.1016/j.future.2020.10.007
Musco C, Musco C (2015) Randomized block Krylov methods for stronger and faster approximate singular value decomposition. In: Cortes C, Lawrence N, Lee D et al (eds) Advances in neural information processing systems, vol 28. Curran Associates, Inc. https://proceedings.neurips.cc/paper/2015/file/1efa39bcaec6f3900149160693694536-Paper.pdf
Nasirigerdeh R, Torkzadehmahani R, Matschinske J et al (2020) sPLINK: a federated, privacy-preserving tool as a robust alternative to meta-analysis in genome-wide association studies. https://doi.org/10.1101/2020.06.05.136382
Nasirigerdeh R, Torkzadehmahani R, Baumbach J et al (2021) On the privacy of federated pipelines. In: International ACM SIGIR conference on research and development in information retrieval 2021. ACM, New York, p 5. https://doi.org/10.1145/3404835.3462996
Pathak MA, Raj B (2011) Efficient protocols for principal eigenvector computation over private data. Trans Data Priv 4(3):129–146
Price AL, Patterson NJ, Plenge RM et al (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38(8):904–909. https://doi.org/10.1038/ng1847
Qi H, Wang TW, Birdwell JD (2003) Global principal component analysis for dimensionality reduction in distributed data mining. In: Statistical data mining and knowledge discovery. Chapman and Hall/CRC, pp 323–338. https://doi.org/10.1201/9780203497159.ch19
Rodríguez MA, Fernández A, Peregrín A et al (2017) A review of distributed data models for learning. Springer, Cham
Ryffel T, Trask A, Dahl M et al (2018) A generic framework for privacy preserving deep learning. arXiv: org/abs/1811.04017
Saad Y (2011) Numerical methods for large eigenvalue problems. Classics in Applied Mathematics, Society for Industrial and Applied Mathematics and Applied Mathematics. https://doi.org/10.1137/1.9781611970739
Sanchez-Fernandez A, Fuente M, Sainz-Palmero G (2015) Fault detection in wastewater treatment plants using distributed PCA methods. In: 2015 IEEE 20th conference on emerging technologies & factory automation (ETFA). IEEE, pp 1–7. https://doi.org/10.1109/ETFA.2015.7301504
Sluciak O, Straková H, Rupp M et al (2016) Distributed Gram–Schmidt orthogonalization with simultaneous elements refinement. EURASIP J Adv Signal Process 1:1–13. https://doi.org/10.1186/s13634-016-0322-6
Steed A, MFD de Oliveira (2010) More than two. Network graphics, pp 125–168. https://doi.org/10.1016/B978-0-12-374423-4.00004-5
Straková H, Gansterer WN, Zemen T (2012) Distributed QR factorization based on randomized algorithms. In: Wyrzykowski R, Dongarra J, Karczewski K et al (eds) Parallel processing and applied mathematics. Springer, Berlin, Heidelberg, pp 235–244
Tam V, Patel N, Turcotte M et al (2019) Benefits and limitations of genome-wide association studies. Nat Rev Genet 20(8):467–484. https://doi.org/10.1038/s41576-019-0127-1
The 1000 Genomes Consortium, Auton, A (2015) A global reference for human genetic variation. Nature 526(7571):68–74. https://doi.org/10.1038/nature15393
Visscher PM, Wray NR, Zhang Q et al (2017) 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet 101(1):5–22. https://doi.org/10.1016/j.ajhg.2017.06.005
Wang S, Chang JM (2019) Differentially private principal component analysis over horizontally partitioned data. In: DSC 2018–2018 IEEE conference on dependable and secure computing, pp 1–8. https://doi.org/10.1109/DESEC.2018.8625131
Wang S, Zhang Z, Zhang T (2015) Improved analyses of the randomized power method and block Lanczos method. arXiv arXiv: org/abs/1508.06429
Wu SX, Wai HT, Li L et al (2018) A review of distributed algorithms for principal component analysis. Proc IEEE 106(8):1321–1340. https://doi.org/10.1109/JPROC.2018.2846568
Funding
Open access funding provided by University Library of Southern Denmark. The FeatureCloud project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 826078. This publication reflects only the authors’ view and the European Commission is not responsible for any use that may be made of the information it contains. AH and DBB were supported by the German Federal Ministry of Education and Research (BMBF, grant no. 031L0309A).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible editor: Pauli Miettinen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hartebrodt, A., Röttger, R. & Blumenthal, D.B. Federated singular value decomposition for high-dimensional data. Data Min Knowl Disc 38, 938–975 (2024). https://doi.org/10.1007/s10618-023-00983-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10618-023-00983-z