Abstract
We discuss topological aspects of cluster analysis and show that inferring the topological structure of a dataset before clustering it can considerably enhance cluster detection: we show that clustering embedding vectors representing the inherent structure of a dataset instead of the observed feature vectors themselves is highly beneficial. To demonstrate, we combine manifold learning method UMAP for inferring the topological structure with density-based clustering method DBSCAN. Synthetic and real data results show that this both simplifies and improves clustering in a diverse set of low- and high-dimensional problems including clusters of varying density and/or entangled shapes. Our approach simplifies clustering because topological pre-processing consistently reduces parameter sensitivity of DBSCAN. Clustering the resulting embeddings with DBSCAN can then even outperform complex methods such as SPECTACL and ClusterGAN. Finally, our investigation suggests that the crucial issue in clustering does not appear to be the nominal dimension of the data or how many irrelevant features it contains, but rather how separable the clusters are in the ambient observation space they are embedded in, which is usually the (high-dimensional) Euclidean space defined by the features of the data. The approach is successful because it performs the cluster analysis after projecting the data into a more suitable space that is optimized for separability, in some sense.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Clustering is the task of uniting similar and separating dissimilar observations in a dataset (Kriegel et al. 2009; Aggarwal 2014). It is a fundamental task in data analysis and is thus widely investigated in many fields. With this study we intend to raise awareness for topological aspects of clustering and to provide empirical evidence that topologically-informed approaches which are conceptually and computationally simple can compete with or even outperform much more complex existing methods on a wide range of problems.
1.1 Problem specification
Cluster analysis is usually approached in an algorithm-driven manner, and considerations about the underlying principles of data generating processes and data structures are often limited to a probabilistic conceptualization assuming that the data X follow a joint probability distribution P(X) (Hastie et al. 2009) or, more precisely, a mixture of distributions (Aggarwal 2014). In contrast, connections to topological data analysis (TDA) (Chazal and Michel 2021; Wasserman 2018), a branch of statistical data analysis inferring the structure of data leveraging topological concepts, are usually not considered. In general, the topological aspects of cluster analysis appear to be an under-investigated topic. Current text books on cluster analysis (Aggarwal and Reddy 2014; Aggarwal 2015; Giordani et al. 2020; Scitovski et al. 2021; Hennig et al. 2015, e.g.) and recent reviews of the field (Jain et al. 1999; Kriegel et al. 2009; Assent 2012; Pandove et al. 2018; Mittal et al. 2019, e.g.) rarely mention the term “topology”.
Following Niyogi et al. (2011), we consider clustering a natural example of TDA. Since an improved understanding of the underlying principles governing the problem is likely to lead to more suitable methods and novel solutions, our work aims to reduce this lack of awareness for topological aspects in the clustering literature. Specifically, our approach follows Niyogi et al (2011, p. 2) who state that “clustering is a kind of topological question” which tries to separate the data into “connected components”. In practice, a connected component is a subset of the vertex set of a graph where the vertices within the subset are connected. For cluster analysis, the nodes of the graph are data points, and the (potentially weighted) edges between them represent their (typically dichotomized or truncated) similarity. Two vertices can be connected by a sequence of connected points, that is, they do not have to be connected directly. One particularly relevant consequence of this topological perspective is its implication that the difficulty of a clustering problem is not necessarily determined by the data’s (nominal) dimensionality.
1.2 Scope of the study
In this work we make use of the well-known algorithm DBSCAN (Ester et al. 1996) for cluster detection and the recently developed manifold learning algorithm UMAP (McInnes et al. 2018) to infer the topological structure of a dataset. To be specific, “inferring the topological structure” as we do here with UMAP has two aspects: first, a fuzzy graph representation of the dataset is used to find the (number of) connected components. Second, this structure is represented by embedding vectors (i.e., coordinates in a representation space) that are optimized for the separability of the connected components. As we show in Sect. 3, UMAP’s graph construction and graph embedding steps both increase cluster separability, and their combined effect thus improves clusterability dramatically. DBSCAN, on the other hand, is a widely used and well established method for cluster detection (Schubert et al. 2017). In particular, it neither requires a pre-specified number of clusters nor does it make any assumptions about their specific shapes or patterns. This is important, as inferring the connected components of a dataset is largely equivalent to identifying the clusters it contains. Moreover, the optimized representation of the topological approach focuses on separability of clusters, not on the specific shapes the clusters might have. Note that UMAP’s developers conjectured that it might enhance density-based clustering, but that this requires further investigation (McInnes 2018).
It needs to be emphasized that that we do not consider UMAP and DBSCAN the most suitable combination in general and our results should not be taken as evidence that this specific combination of methods is the most suitable for the purpose. Other combinations of clustering and/or manifold learning methods than UMAP and DBSCAN are possible, can lead to comparable or even better results, and certainly deserve investigation as well. For example, note the results of Allaoui et al. (2020) that show that combining UMAP with k-means can yield comparable results. That said, this work is not intended as a benchmark study comparing clustering approaches.
Instead, the main contribution of the work is to demonstrate that inferring the topological structure of a dataset before attempting to partition its clusters can considerably enhance the resulting partition and how easily it can be found, and we use the specific combination of UMAP and DBSCAN to demonstrate this. UMAP is used to preprocess the data such that its representation is optimized for separability and the resulting embedding vectors are used as inputs for DBSCAN.
We chose UMAP in particular as our embedding method for the following reasons. First of all, UMAP has a decidedly topological underpinning, so it is suitable for a theoretical analysis from the clustering perspective we take here. In particular, it builds on simplicial complexes to obtain a fuzzy topological representation of the inherent structure of a dataset. As such, it is based on the same theoretical principles as topological data analysis (Chazal and Michel 2021; Wasserman 2018). These aspects are outlined in more detail in Sect. 2.2. In addition, it has already been shown that preprocessing by UMAP can improve clustering results (Allaoui et al. 2020) and, moreover, that the resulting embeddings frequently yield “more compact clusters than t-SNE [another state-of-the-art manifold learning method] with more white space in between” (Kobak and Linderman 2021, p. 157). The topological embedding methods PAGA (Wolf et al. 2019) and Topomap (Doraiswamy et al. 2021) are domain-specific and/or by far not as well-established and generally applicable as UMAP. Other alternatives to infer topological structure from data are not suited for pre-processing as needed here. For example, persistent homology diagrams (Wasserman 2018) do not provide a vector representation of the dataset that could be supplied as input to a clustering method, and neither do measures of data separability such as those described in Guan and Loew (2021), which only yield scalar values indicating class overlap.
We again emphasize that the goal of the study is to describe and explain why clustering embedding vectors representing the inherent topological structure of a dataset can be highly beneficial, and we use the specific combination of UMAP and DBSCAN in our experiments to practically demonstrate this. The aspects outlined above provide reasons to select these methods, but we do not indent to assert and establish the superiority of this particular combination of methods in general. As outlined, other combinations of clustering and/or manifold learning methods are possible and may lead to comparable or even better results.
1.3 Contributions
This study makes three distinct contributions: First, Sect. 3 illustrates that approaches motivated by a topological perspective can dramatically reduce the difficulty of clustering for both low- and high-dimensional data. This is achieved with an in-depth analysis of simulated data specifically designed to reflect some often described problems of clustering including high-dimensional data, clusters of different density, and irrelevant features. In addition, a simple toy example demonstrates why and how inferring the intrinsic topological structure of a dataset with UMAP before clustering improves the clustering performance of DBSCAN.
Secondly, with intuition and motivation in place, Sect. 4 is devoted to specific implications of the topological perspective. We describe which structures of a dataset are preserved when inferring the topological structure by finding connected components and enhancing separability (using the UMAP algorithm), in particular by contrasting topological against geometrical characteristics in a detailed qualitative and quantitative analysis of simple synthetic examples.
Finally, in Sect. 5, we report extensive experiments using real world data. Our results show that inferring the topological structure of datasets before clustering them not only improves—dramatically, for some examples such as MNIST—performance of DBSCAN, but also drastically reduces its parameter sensitivity. The comparatively simple approach of combining UMAP and DBSCAN can even outperform recently proposed clustering methods such as ClusterGAN (Mukherjee et al. 2019), which require expensive hyperparameter tuning, on complex datasets.
In addition, related work and the methods used are described in Sect. 2, while the results are discussed in Sect. 6 before we conclude in Sect. 7.
2 Methods and related work
In this section, we first describe the background of the study and related work, before we outline the methods DBSCAN and UMAP, which are used for clustering and inferring topological structure, respectively, in this study. Readers which are familiar with the methods might skip the corresponding paragraphs. However, note that we will refer to some of the more technical details outlined here in Sect. 3.2.
2.1 Background and related work
The body of literature on clustering, topological data analysis and manifold learning is extensive and has seen contributions from many different areas and perspectives. General reviews on clustering have been provided for example by Jain et al. (1999) and more recently by Saxena et al. (2017). Moreover, there a several reviews focusing on cluster analysis for high-dimensional data (Kriegel et al. 2009; Assent 2012; Pandove et al. 2018; Mittal et al. 2019). In addition, there exist overviews on TDA (Niyogi et al. 2011; Chazal and Michel 2021; Wasserman 2018, e.g.) as well as on manifold and representation learning (Cayton 2005; Bengio et al. 2013; Wang et al. 2021) including the textbooks by Ma and Fu (2012) and Lee and Verleysen (2007).
The variety of clustering algorithms is vast and endeavors have been made to capture this diversity through taxonomies. DBSCAN, the algorithm used here, is a density-based approach. One of its major advantages is that it does not require a pre-specified number of clusters and that the clusters can have arbitrary shapes and patterns. Its hierarchical version (HDBSCAN, Campello et al. 2013) does not use a global \(\varepsilon\)-threshold but computes on its own multiple cut-off values resulting in clusters of different densities and therefore requires only the minPts parameter. Similar to HDBSCAN, the OPTICS algorithm (Ankerst et al. 1999) calculates an ordering of the observations without a global \(\varepsilon\)-threshold that provides broader insight on the structure of the data. However, the method does not explicitly assign cluster memberships. Instead, it allows to visualize the hierarchical cluster structure for example via reachability plots (Ankerst et al. 1999).
Further categories are hierarchical and partitioning algorithms (Jain et al. 1999), where the latter can be divided further into sub-taxonomies. Some of them are based on the minimization of distances to certain prototypes (centroids, medoids, etc.), this includes algorithms like k-means (Lloyd 1982), or its more general archetype of algorithms: Gaussian Mixture Models (GMMs) among which the Expectation-Maximization (EM) algorithm (Dempster et al. 1977) is a prominent exponent. A major caveat, however, is that these methods estimate a specific probabilistic model which includes the number of clusters to be detected and often fail if the data is distributed differently (Liu and Han 2014). Note, however, that there are approaches for determining the number of clusters (Hamerly and Elkan 2003; Pham et al. 2005; Mehar et al. 2013; Debatty et al. 2014, e.g.).
In contrast, spectral clustering, a family of algorithms that shares some common ground with many manifold learning methods, are more robust with respect to shape and distribution of the clusters. However, note that these methods require the number of clusters to be specified in advance (Von Luxburg 2007; Liu and Han 2014). They compute the spectral decomposition of the Laplacian of a graph obtained from a pairwise (dis)similarity matrix and there are several results that show that such low-rank matrix approximations can improve cluster separability. Blum et al (2020, Ch. 7.5), for example, prove that cluster separability can be increased if one computes a singular value decomposition of the data matrix. In a similar vein, Cohen-Addad and Schwiegelshohn (2017) study distribution stability, perturbation resilience, and—in particular—spectral separability, three conditions that allow to distinguish different data settings and can simplify clustering.
Subspace clustering approaches emerged specifically for high-dimensional settings (Kriegel et al. 2009; Assent 2012; Pandove et al. 2018; Mittal et al. 2019). The fundamental assumption here is that objects within a cluster do not exhibit high similarities among all dimensions but only within a small subset of features that can either (a) span an axis-parallel subspace or (b) an affine projection to an arbitrarily-oriented subspace (“correlation clustering”). In both cases, the objects of a cluster are assumed to be located on a common, low-dimensional linear manifold.
In contrast, manifold learning is based on the assumption that data observed in a high-dimensional ambient observation space is distributed on or near a potentially nonlinear manifold with a much smaller intrinsic dimension than the ambient space (Ma and Fu 2012). In general, the aim is to find low-dimensional representations of datasets preserving as much of the structure of the observed data as possible. A synonymous term is nonlinear dimension reduction (NDR) (Lee and Verleysen 2007). However, there is no general definition of which characteristics are to be preserved and represented and different methods infer the intrinsic structure and provide low-dimensional representations in different ways.
For instance, principal component analysis (PCA) yields embedding vectors that optimally preserve global Euclidean distances in the original data space, while other methods such as Isomap (Tenenbaum et al. 2000) yield embedding vectors that aim to preserve geodesic distances on a single, globally connected data manifold. Methods like t-distributed Stochastic Neighbor Embedding (t-SNE, van der Maaten and Hinton 2008) and uniform manifold approximation and projection (UMAP, McInnes et al. 2018) have been successfully applied to complex high-dimensional datasets with cluster structure. More recently, methods with a specific topological focus such as general purpose Topomap (Doraiswamy et al. 2021) as well as domain specific Paga (Wolf et al. 2019), which focuses on the analysis of single cell data, have been proposed. The manifold learning-based clustering approach of Souvenir and Pless (2005) relies on the assumption that data is sampled from multiple intersecting lower-dimensional manifolds.
Modern manifold learning methods like UMAP and t-SNE optimize a loss functions—the cross-entropy in UMAP, the Kullback–Leibler divergence in t-SNE—via (stochastic) gradient descent (Wang et al. 2021, e.g.). Therefore, an initial configuration of points is required to start the optimization procedure. The initialization procedure has a crucial effect on the embedding performance and, in general, a non-random initialization should be used (Kobak and Linderman 2021; Wang et al. 2021; McInnes et al. 2018, e.g.). UMAP uses a spectral embedding initialization by default. Other methods, for example, TriMAP and PaCMAP, use PCA for initialization (Wang et al. 2021). t-SNE, in contrast, uses a random initialization by default in many implementations but should also be initialized non-randomly by methods like PCA (Kobak and Linderman 2021). In summary, these methods leverage standard spectral embedding methods such as PCA or Laplacian Eigenmaps but additionally make use of more sophisticated graph construction and embedding layout steps that improve the embedding performance. This also illustrates the close connections between manifold learning and spectral clustering. As outlined, spectral clustering uses a (dis)similarity matrix (and the number of clusters \(n_c\)) as input. From that, it first constructs a similarity graph and then computes the spectral decomposition of the corresponding graph Laplacian that encodes the graph structure. Using the first \(n_c\) eigenvectors as inputs to the k-means algorithm finally yields the desired clustering (Von Luxburg 2007). More generally, one can conduct spectral clustering directly using a \(n \times d\) data matrix X instead of a \(n \times n\) (dis)similarity matrix by computing the singular value decomposition (SVD) of X (Blum et al. 2020, Ch. 7.5). Note that this (lower rank) matrix factorization is a projection to a lower dimensional subspace of dimension \(n_c\) that can increase cluster separability because it “brings data points closer to their cluster centers” (Blum et al. 2020, p. 222). For more on the connections between manifold learning and spectral clustering see, for example, Belkin and Niyogi (2001), Saul et al. (2006) and Linderman and Steinerberger (2019).
Several studies that precede ours also focus on the combination of manifold learning techniques and cluster analysis, with applications to cytometry data (Putri et al. 2019), brain tumor segmentation (Kaya et al. 2017), spectral clustering (Arias-Castro et al. 2017), or big data (Feldman et al. 2020), the latter three based on PCA. DBSCAN was used in combination with multidimensional scaling (MDS) in Mu et al. (2020), and UMAP was used for time-series clustering (Pealat et al. 2021) as well as clustering SARS-COV-2 mutation datasets (Hozumi et al. 2021). However, these all focus on specific domains and not on the underlying topological principles. In contrast, we base our work on a topological perspective on clustering first described theoretically by Niyogi et al. (2011), who conceptualize clustering as the problem of identifying the connected components of a data manifold. We show the theoretical and practical utility of this perspective by means of extensive experiments based on synthetic and real datasets. Similar in spirit to our work, Allaoui et al. (2020) perform a comparative study with real data to show that UMAP can considerably improve the performance of clustering algorithms. Among other things, they combined UMAP with HDBSCAN and report comparable clustering results for three of the real-world datasets (Pendigits, MNIST and FMNIST) also used here. However, in contrast to our study, Allaoui et al. (2020) do not provide insights into the conceptual topological underpinnings, nor do they describe how the data structures preserved in UMAP embeddings lead to these performance improvements. Note that their results also show empirically that the benefits of the proposed approach are not tied to any particular combination of NDR and clustering methods.
2.2 UMAP
Three important assumptions (or “axioms”) underlie the method UMAP. It is assumed that “there is a manifold on which the data would be uniformly distributed”, “the underlying manifold of interest is locally connected”, and “preserving the topological structure [...] is the primary goal” (McInnes et al. 2018, p. 13). UMAP is rather similar to other manifold learning methods in its basic computational structure, and we first outline these aspects of UMAP in the following. We then try to build up some intuition about the theoretical underpinnings. The principle idea behind UMAP essentially consists of two steps:
-
1.
Constructing a weighted \(k\)-nearest neighbor (\(k\)-NN) graph from a pairwise distance matrix.
-
2.
Finding a (low-dimensional) representation of the graph which preserves as much of its structure as possible.
Note that this is the fundamental principle in manifold learning and the details of the two steps constitute the differences between manifold learning methods (Wang et al. 2021). However, unlike many other manifold learning methods, UMAP is based on a solid theoretical foundation that ensures that the topology of the manifold is faithfully approximated (McInnes et al. 2018)
2.2.1 Graph construction
Given a dataset \(X = \{x_1,..., x_{n_{{\text {obs}}}}\}\) sampled from a space equipped with a distance metric \(d(x_i, x_j)\), UMAP constructs a directed \(k\)-NN graph \(\bar{G} = (V, E, w)\) with the vertices \(V_i\) being observations \(x_i\) from X, E the edges and w the weights, based on the following definitions.
Definition 1
The distance \(\rho _i\) of an observation \(x_i\) to its nearest neighbor is defined by
Definition 2
A (smooth) normalization factor \(\sigma _i\) is set for each \(x_i\) by
This defines a local (Riemannian) metric at point \(x_i\).
Definition 3
Weight function: The edge weights of the graph are defined by
Note that w takes on values in the interval [0, 1]. Moreover, the distance to the nearest neighbor \(\rho _i\) ensures that \(x_i\) is connected to at least one other point with an edge of weight 1 (local connectivity constraint).
For the theory to work it is essential to assume that the data is uniformly distributed on the manifold, which is too strong an assumption for real world data. The issue is bypassed by defining independent notions of distance at each observed point through \(\sigma _i\) and \(\rho _i\). However, these local metrics may not be mutually interchangeable, which means that the “distance” between neighboring points \(x_i\) and \(x_j\) may not be the same if measured w.r.t \(x_i\) or w.r.t. \(x_j\), i.e., \(d(x_i, x_j) \ne d(x_i, x_j)\), so edge weights in \(\bar{G}\) depend on the direction of the edges.
A unified, undirected graph G with adjacency matrix B is obtained by
with A the weighted adjacency matrix of \(\bar{G}\) and \(\circ\) the Hadamard productFootnote 1. Note that Eq. (1) represents the well defined operation of unioning fuzzy simplicial sets (with which the manifold is approximated). The resulting entries in B can be interpreted as the probability that at least one of the two directed edges between two vertices in \(\bar{G}\) exists, or more generally as a measure of similarity between two observations \(x_i\) and \(x_j\). Note that it has recently been shown that a stricter notion of connectivity induced by mutual nearest neighbors can further improve the topology preserving property of standard UMAP used here (Dalmia and Sia 2021). However, at the time of writing this paper, no implementation of this approach was publicly available.
2.2.2 Graph embedding
The objective is to find a configuration of points in the representation space Y whose fuzzy simplicial set is as similar as possible to the fuzzy simplicial set of the original data, as represented by G. To find this low-dimensional representation, UMAP optimizes the cross entropy of edge weights in the two spaces. Similarities in the observation space are represented in terms of the local smooth nearest neighbor distances as
with \(v_{j \vert i} = \exp [(-d(x_i, x_j) - \rho _i)/\sigma _i]\) (c.f. Eq. (1)), and similarities in the representation space Y as
with \(y_i\) and \(y_j\) the embedding vectors to optimize and a and b tuning parameters that are—by default—\(a \approx 1.929\) and \(b \approx 0.7915\). The cross entropy between the two fuzzy simplicial set representations
is minimized via stochastic gradient descent (SGD) to obtain the graph layout. The two terms in Eq. (4) represent the attractive and repulsive forces for the graph layout algorithm used here.
UMAP’s central tuning parameters are the number of nearest neighbors \(k\) (often denoted as n or n_neighbors), the number of SGD optimisation iterations n-epochs, the dimension d of the representation space, and min-dist, a parameter controlling how close neighboring points can appear in the representation.
2.2.3 Topological background
In the following, we try to build up some intuition about UMAP’s theoretical underpinnings by describing its close connections to topological data analysis (TDA). Note that UMAP has a sophisticated category theoretical foundation, but in principle the basic conceptual and computational cornerstones are simplicial complexes, sets of \(n\)-simplices that can be constructed using the \(n+ 1\) points in a given set. In more descriptive terms, 0-simplices are points, 1-simplices are edges between points, 2-simplices are triangles, 3-simplices are tetrahedrons, and so on. A subset of a \(n\)-simplex is called a face and is a (lower order) simplex itself (Wasserman 2018; Zomorodian and Carlsson 2005).
More generally, given a set \({\mathcal {X}}\), the vertex set, an abstract simplicial complex can be defined as the “set \({\tilde{K}}\) of finite subsets of \({\mathcal {X}}\) such that the elements of \({\mathcal {X}}\) belong to \({\tilde{K}}\) and for any \({\textbf{s}}\in {\tilde{K}}\), any subset of \({\textbf{s}}\) belongs to \({\tilde{K}}\)” (Chazal and Michel 2021, p. 4). Chazal and Michel (2021, p. 4) emphasize that “abstract simplicial complexes can be seen as topological spaces”. That means, if a simplicial complex is constructed from a given data set \(X = \{x_1,..., x_{n_{{\text {obs}}}}\}\), one obtains a topological representation of the the dataset, encoding information on structures such as connected components (i.e., clusters) or holes. On the other hand, simplicial complexes are combinatorial objects and thus allow for efficient computations (Chazal and Michel 2021).
In practice, one can, for example, compute a Vietoris-Rips complex \(VR_r(X)\) (Wasserman 2018; Chazal and Michel 2021). Given a dataset \(X \subset {\mathbb {R}}^D\) (note that \({\mathbb {R}}^D\) is a metric space naturally endowed with the Euclidean metric) and \(r \in {\mathbb {R}}^+_0\), a Vietoris-Rips complex is the set of simplices with \(d(x_i, x_j) \le r\), \(x_i, x_j \in X\). The value r controls the resolution with which the topological features are extracted from the (finite) dataset and each point will appear as one of \(n_{{\text {obs}}}\) unconnected components if \(r = 0\). On the other extreme, a single connected component will result for a large r, i.e., all observations will appear connected to each other (Bubenik 2015).
Computing filtrations, that is, a series of Vietoris-Rips complexes for increasing values of r, is a very important approach in TDA called persistent homology (Chazal and Michel 2021; Zomorodian and Carlsson 2005, e.g.), where “homology characterizes sets based on connected components and holes” (Wasserman 2018, p. 515). That means, starting from the observations in a dataset as \(n_{{\text {obs}}}\) unconnected components, connected components and other topological features such as holes or voids will appear and vanish with increasing r (Wasserman 2018). The birth and death times of these topological features can be used to create a persistence diagram, with the birth time plotted on the horizontal axis and the death time plotted on the vertical axis. Persistent topological features, i.e., features with long lifetimes, appear far from the diagonal.
UMAP, in contrast, constructs a single simplicial complex in its graph construction step in principle. The simplicial complex is constructed by specifying a number of nearest neighbors k to include in a ball around a point. This results in locally different metrics because in non-uniformly distributed data the distance to the kth neighbor depends on how densely sampled the region is a point is located in. In more descriptive terms, consider that the radius of the ball around a point \(x_i\) is equal to the distance to \(x_i\)’s k-nearest neighbor. Since k is the same for all points, the radii of balls around points in dense regions are smaller than those of points in sparse regions (McInnes 2018). In particular, as outlined, each edge can have two different weights depending on the point from which the distance is measured. In the more abstract terms on which UMAP is built, this means that one obtains a family of fuzzy simplicial sets. A simplicial set is a more general notion of a simplicial complex usually defined in category theoretical terms (McInnes et al. 2018). In particular, the vertices of a simplicial set “need not be distinct nor necessarily determine the simplex spanning them” (Riehl 2011, p. 2). The simplicial sets are fuzzy because of the two different weights an edge can obtain and computing the fuzzy union of the fuzzy simplicial sets results in a single weight for each edge (McInnes 2018).
In summary, both TDA and UMAP approximate continuous structures by building simplicial complexes and sets from the (discretely) observed data points. UMAP constructs a topological representation of a dataset via fuzzy simplicial sets while persistent homology, as a specific example of TDA, yields a persistence diagram that indicates topological features of the data based on filtrations. These close connections between TDA and UMAP emphasize UMAP’s topological “nature”. If the data live on a manifold (a single connected component), UMAP constructs a topological representation of the manifold. On the other hand, if the data live on different unconnected components, UMAP constructs a representation that preserves these unconnected components. In particular, as will be demonstrated in Sect. 3.2, if the parameter k steering the neighborhood size is much smaller than the number of observations \(n_{{\text {obs}}}\), many edge weights will become 0 in the graph construction step with the local connectivity constraint (see Definition 3) ensuring that the dataset is not separated into many connected components consisting of a single point only. In general, UMAP should not be considered an isometric embedding method, i.e., a method preserving distances, but rather a method preserving topological features.
2.3 DBSCAN
Since DBSCAN is already very well-known and established, we will only briefly describe its principles. The idea behind DBSCAN is captured within six definitions we adapt from Ester et al. (1996) and elaborate on:
Definition 4
\(\varepsilon\)-neighborhood of an object: The \(\varepsilon\)-neighborhood of an object \(x_i\) denoted by \({\mathcal {N}}_{\varepsilon }(x_i)\), is defined by:
where X denotes a given dataset.
Definition 5
Directly density-reachable: An object \(x_i\) is directly density-reachable from an object \(x_j\) w.r.t. a given \(\varepsilon\)-range and MinPts if:
-
1.
\(x_i \in {\mathcal {N}}_{\varepsilon }(x_j)\) and
-
2.
\(\vert {\mathcal {N}}_{\varepsilon }(x_j) \vert \ge MinPts\) (core point condition)
Definition 6
Density-reachable: An object \(x_i\) is density-reachable from another object \(x_j\) w.r.t. \(\varepsilon\) and MinPts if there is a chain of objects \(x_1,...,x_c\), \(x_1 = x_i\), \(x_c = x_j\) such that \(x_{l+1}\) is directly density-reachable from \(x_l\).
Definition 7
Density-connected: An object \(x_i\) is density-connected to another object \(x_j\) w.r.t. \(\varepsilon\) and MinPts if there is an object o such that both, \(x_i\) and \(x_j\) are density-reachable from o w.r.t. \(\varepsilon\) and MinPts.
Definition 8
Cluster: Let X be a given dataset of objects. A cluster C w.r.t. \(\varepsilon\) and MinPts is a non-empty subset of X satisfying the following conditions:
-
1.
\(\forall x_i,x_j:\) if \(x_i\in C\) and \(x_j\) is density-reachable from \(x_i\) w.r.t. \(\varepsilon\) and MinPts, then \(x_j \in C\) (Maximality)
-
2.
\(\forall x_i,x_j \in C: x_i\) is density-connected to \(x_j\) w.r.t. \(\varepsilon\) and MinPts (Connectivity)
Definition 9
Noise: Let \(C_1,...,C_{n_c}\) be the \(n_c\) clusters of the given dataset X w.r.t. parameters \(\varepsilon _i\) and \(MinPts_i\), \(i=1,...,n_c\). Then noise is defined as the set of objects in the dataset X that do not belong to any cluster \(C_i\), i.e. \(noise = \lbrace x_i \in X \vert \forall i: x_i \notin C_i \rbrace\).
In Definition 5 an object is a core point if it has at least MinPts number of objects within its \(\varepsilon\)-neighborhood. In the case that no objects in a given dataset are density-reachable then we would obtain \(n_c\) clusters where \(n_c\) denotes the number of core-points in a dataset X for a given \(\varepsilon\) and MinPts. This means that the number of core points can be considered as an upper-bound for the number of emerging clusters for a given \(\varepsilon\) and MinPts. Further it can be deduced from the core point definition that the region surrounding a core point is more dense compared to density-connected objects that do not satisfy \(\vert {\mathcal {N}}_{\varepsilon } (x_j) \vert \ge MinPts\) meaning that they are objects in more sparse regions.
3 Inferring the topological structure enhances clusterability
In this section, we demonstrate that the correct use of manifold learning (here, specifically: UMAP), as motivated by our topological framing, largely avoids several frequently described challenges in cluster analysis.
A major problem affecting cluster analysis is that clustering often becomes more challenging in high-dimensional datasets. Specifically, the presence of many irrelevant and/or dependent features potentially degrades results (Kriegel et al. 2009). However, contrary to wide-spread “folk-methodological” superstitions and some sources like Assent (2012), the well known result that \(L_p\)Footnote 2 distances lose their discriminating power in high dimensions (Beyer et al. 1999, e.g.) is entirely irrelevant for well-posed clustering problems: both the original publication and subsequent works like Kriegel et al. (2009) and Zimek and Vreeken (2015) show that the conditions for this result do not apply if the data is distributed in well separable clusters. In particular, this means that DBSCAN, being based on pairwise distance information, can easily detect clusters in high-dimensional datasets.
Nevertheless, there are other problems specific to density-based clustering, and DBSCAN in particular, among which finding a suitable density level is one of the most important (Kriegel et al. 2011; Assent 2012). A recent review (Schubert et al. 2017), outlined some heuristic rules for specifying \(\varepsilon\) for DBSCAN, but domain knowledge should mostly determine such decisions. More importantly, density based clustering is likely to fail for clusters with varying density. In such cases, a single global density level—for example, specified via \(\varepsilon\) in DBSCAN—cannot delineate cluster boundaries successfully (Kriegel et al. 2011).
In addition to these well known issues, we outline another more subtle, less well known aspect: not only does the difficulty of a clustering problem not necessarily increase for high-dimensional X, clusters may even become easier to detect in higher dimensional (embedding) spaces.
3.1 Enhancing clusterability of DBSCAN with UMAP
The four example datasets we consider here illustrate the following three points: (1) Density-based clustering works in some but not all high-dimensional settings. (2) Perfect performance may not be achievable even for extensive parameter grid searches, and suitable \(\varepsilon\) values are highly problem-specific. (3) Most importantly, manifold learning can considerably enhance clustering both by improving performance and by reducing parameter sensitivity of DBSCAN to the extent it becomes almost tuning-free.
The datasets we consider here consist of three clusters sampled from three multivariate Gaussian distributions with different mean vectors. In the first two examples, denoted by \(E_{100}\) and \(E_{1000}\), the covariance matrix for all three Gaussians is the identity matrix, inducing clusters of similar density. In the latter two examples, \(U_3\) and \(U_{1003}\), the covariance matrices differ, inducing clusters of different density. In addition, we consider problems with very different dimensionalities. Observations in setting \(E_{100}\) are sampled from 100-dimensional Gaussians, while observations in setting \(E_{1000}\) are sampled from 1000-dimensional Gaussians. In contrast, observations for \(U_3\) and \(U_{1003}\) are sampled from 3-dimensional Gaussians. For \(U_{1003}\), an additional 1000 features that are irrelevant for cluster membership are sampled independently and uniformly from [0, 1]. For each setting we sample 500 observations from each of the three clusters, i.e., each example dataset consists of 1500 observations in total. The complete specifications of the examples are given in Table 1.
We use the Adjusted Rand Index (ARI) (Hubert and Arabie 1985, Eq. 5) and the Normalized Mutual Information (NMI) with maximum normalization (Vinh et al. 2010a, Tab. 2) as (quantitative) evaluation measures throughout. Both measures compare two data partitions and return a numeric value quantifying the agreement. The NMI strictly ranges between [0, 1], but it is not adjusted for random partition. The ARI also takes values in the range [0, 1], but unlike the NMI, it is adjusted for random partitioning. Strictly speaking, this means the ARI becomes 0 only when the Rand Index exactly matches its expected value under the null hypothesis that the partitions are generated randomly from a hypergeometric distribution (Hubert and Arabie 1985).
That means, we focus on external cluster evaluation in this work, a frequently used approach in methodological research on cluster analysis. Note, however, that in truly unsupervised and exploratory settings relevant external information is often unavailable. In such situations, one can fall back on internal evaluation measures to evaluate and choose between clusterings obtained with different parameter settings in practice. Unlike external evaluation measures that evaluate a cluster result according to its concordance with the ground-truth partition (synthetic data) or a label set (real data), internal evaluation measures evaluate a clustering based on an objective such as “within-cluster homogeneity, between-cluster separation, and similarity of cluster members to their cluster centroid” (Van Mechelen et al. 2018, p. 15). Prominent examples of internal evaluation measures are the silhouette coefficient (Rousseeuw 1987), Hopkins statistics (Hopkins and Skellam 1954), and the Davis-Bouldin-Index (Davies and Bouldin 1979). Fig. 1 shows the ARI and NMI for different \(\varepsilon\) values obtained by either applying DBSCAN directly to the observed data or to their 2D UMAP embeddings. Note that we refer, e.g., to a 2-dimensional UMAP embedding obtained with \(k = 5\) as 2D UMAP with \(k = 5\) or 2D UMAP-5 for short.
Several aspects need to be emphasized. First of all, the effect of the dimensionality of the dataset on the performance of DBSCAN applied to the original data is complicated (Fig. 1, first column (A)). Contrary to preconceived notions, it can be easier to detect clusters in higher dimensions. Figure 1A shows that using only DBSCAN, clusters are more easily detected in the 1000-dimensional data (2nd row) than in the 100-dimensional data (1st row, although perfect performance is not achieved by DBSCAN in either of the two).
ARI and NMI as a function of \(\varepsilon\) for the synthetic settings \(E_{100}, E_{1000}, U_3, U_{1003}\). First column: DBSCAN directly applied to the data. Second column: DBSCAN applied to a 2D UMAP embedding with \(k= 5\). Clusters sampled from multivariate Gaussian distributions (see Table 1 for specifications). For setting \(U_{1003}\), additional 1000 irrelevant variables are sampled uniformly from [0, 1]. DBSCAN computed for \(\varepsilon \in [0.01, 50]\), step size: 0.01; \(minPts = 5\)
The dimension of the Gaussian distributions defining the clusters is the only difference between these two settings. On the other hand, Fig. 1A shows that it can also be the other way round. In the 1003-dimensional dataset with 1000 irrelevant features (4th row) cluster performance is much lower than in the corresponding 3-dimensional dataset with only 3 relevant variables (3rd row). Again, perfect cluster performance is not achieved by DBSCAN alone. Note that settings \(U_3\) and \(U_{1003}\) define clusters with varying densities, so DBSCAN is expected not to provide a perfect result.
Secondly, finding a suitable value of \(\varepsilon\) is very challenging using DBSCAN alone. Note that the optimal \(\varepsilon _{opt}\) varies between 0.9 and 42.64 for these examples. Identifying a suitable \(\varepsilon\) is even more problematic since the sensible \(\varepsilon\)-ranges are very small (e.g. see \(U_{1003}\)). In some cases, clustering does not seem feasible at all even with an optimally chosen \(\varepsilon\)—optimal results are very poor for setting \(E_{100}\) with ARI (NMI) \(= 0.003 (0.05)\) for \(\varepsilon _{opt} = 11.32 (10.98)\). Moreover, while \(\varepsilon _{opt}\) is not necessarily consistent for datasets with approximately the same dimensionality—compare \(\varepsilon _{opt} = 42.64\) for \(E_{1000}\) to \(\varepsilon _{opt} = 12.48\) for \(U_{1003}\)—it can be similar for datasets with very different dimensionality—compare \(\varepsilon _{opt} \sim 11\) for \(E_{100}\) to \(\varepsilon _{opt} = 12.48\) for \(U_{1003}\).
Finally, the crucial point we want to highlight with these examples is that inferring the topological structure before clustering by applying DBSCAN on UMAP embeddings instead of directly to the data makes all these issues (almost) completely disappear (see Fig. 1B). First of all, clustering performance is increased in all four examples; in three it even leads to perfect performances. But not only is performance increased, UMAP also dramatically reduces the complexity of finding a suitable \(\varepsilon\). In all considered cases the sensible \(\varepsilon\)-ranges start near zero, rapidly reach the optimal value, and remains optimal over a wide range of \(\varepsilon\)-values in three of the four examples. Note that we do not tune UMAP at all – we simply set \(k= 5\) and leave all other settings at their default values.
We emphasize that perfect performance is obtained for large swaths of the \(\varepsilon\)-range we consider for the two high-dimensional examples. This suggests that the crucial issue in clustering is not the nominal dimension of the dataset or whether it contains irrelevant features, but rather how separable the clusters are in their ambient space, which is usually simply the p-dimensional Euclidean space spanned/defined by the dimensions/features of the data, while the approach taken here attempts to cluster observations after projecting them into a space that is optimized for separability.
To analyze the variability across different samples, we generated another 50 datasets for each of the settings (as defined in Table 1) and report the means and standard deviations for ARI and NMI in Table 2. For these experiments, we set \(\epsilon = \epsilon _{opt}\) (the values corresponding to the optimal ARI values shown in Fig. 1), i.e., for DBSCAN applied directly to the data, \(\epsilon = \{11.32, 42.64, 0.9, 12.48\}\), and for DBSCAN applied to 2D UMAP-5 embeddings, \(\epsilon = \{0.24, 0.46, 1.41, 0.28\}\). The results show that the findings are very robust across different samples. In particular, the mean values are close to the optimal values shown in Fig. 1 with (very) low variability. See “Appendix A” for a more details.
In summary, applying DBSCAN on UMAP embeddings not only improved performance considerably, it also reduced the sensitivity of DBSCAN w.r.t. \(\varepsilon\). In particular, suitable \(\varepsilon\)-ranges started near zero for all considered examples. Our experiments described in Sect. 5 show that this holds for complex real data such as fashion MNIST (Xiao et al. 2017) as well, where applying DBSCAN on UMAP embeddings not only dramatically improved DBSCAN’s performance but even outperformed the recently proposed ClusterGAN (Mukherjee et al. 2019) method. In the next subsection, we examine the technical aspects that explain this behavior in a simple toy example.
3.2 Reasons for improved clusterability
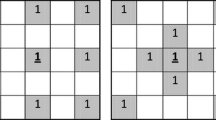
This section lays out possible reasons for the observed improvements w.r.t clusterability with a detailed analysis of the underlying technical mechanisms in a simple toy example. Consider the following distance matrix between six objects:

Inspecting this distance matrix reveals two clusters of objects, shown here in green and cyan. We set DBSCAN’s core point condition parameter to \(minPts = 2\). Note that the object itself is not considered part of its \(\epsilon\)-neighborhood. We set \(\varepsilon = 0.75\), so that every object whose row (or column) in the distance matrix contains at least two entries \(\le 0.75\) is considered a “core point”. Since two objects from the different clusters have a distance of exactly 0.75 (orange entries), all objects are part of a single connected component, and the two dense regions are subsumed into a single large cluster for \(\varepsilon = 0.75\), as can be seen in the matrix below:

To avoid this collapsed solution, one could try to reduce the \(\varepsilon\) parameter to e.g. \(\varepsilon = 0.74\). However, as a consequence, now all the objects in the second (cyan) cluster become “noise”: They no longer satisfy the “core point” condition for \(minPts = 2\), since at most one distance in each of their rows is \(\le 0.74\). This means only one cluster (top left, green) is detected, as can be seen in the following matrix:

From this first example, we conclude (1) that there may be cases where even a single object may connect two clusters, yielding a single collapsed cluster and (2) that the sensitivity of clustering solutions to hyperparameter settings is large: A small change of the \(\varepsilon\)-parameter by only 0.01 led to a fundamentally different solution.
Thus, we should look for improvements that (i) reduce the sensitivity of results towards the parameter settings and (ii) increase the separability of the data and thereby reduce the susceptibility of DBSCAN to merge multiple poorly separated clusters via interconnecting observations at their respective margins. Sharpening the distinction between dense and sparse regions within the dataset, i.e. increasing separability, improves clusterability. As we will now see, UMAP is able to do exactly that by arranging objects into clusters with fairly constant density within and empty regions in between.
To illustrate this, we consider the representation of the toy example via the fuzzy graph as constructed by UMAP. This reflects the fuzzy simplicial set representation of the data and crucially depends on the number of nearest neighbors \(k\). We start with \(k= 6\). This leads to a graph with adjacency matrix
Each cell represents the fuzzy edge weight \(v_{ij}\) (Eq. 2) connecting two points, so each value represents the affinity of two observations, not their dissimilarity as in the distance matrices before. As before, the cluster structure is obvious in this representation, with high affinities (\(\ge 0.95\)) where distances had been low (\(\le 0.75\)). The representation learned by UMAP in the graph construction step clearly reflects the cluster structure of the dataset.
Note that this fuzzy topological representation by itself already amplifies the cluster structure: if we stopped UMAP at this point and converted the affinities \(v_{ij}\) into dissimilarities e.g. via \(d_{ij} = 1 - v_{ij},\) \(i \ne j\), DBSCAN with \(minPts = 2\) would yield perfect cluster results for \(\varepsilon \in [0.01, 0.09]\)!
Note as well that UMAP’s graph layout optimization has not even been performed yet and that the nearest-neighbor parameter \(k\) has been set to 6, the largest possible value in this example. Thus, the vast improvement in separability we observe is due only to the way UMAP learns and represents the structure of the data in the fuzzy graph G alone. The improvement can be driven even further both by decreasing the parameter \(k\) and by conducting the graph layout optimization.
First, consider the effect of \(k\). In the following, blanks in the matrices denote zero entries. Graph (9) shows G for \(k= 3\). Clearly, the beneficial effects we noted for \(k= 6\) are considerably amplified.
Almost all \(v_{ij}\) become zero (i.e. there is no affinity/similarity between the two points) except for those joined in one of the clusters and the two entries which caused DBSCAN to break. Turning \(v_{ij}\) into \(d_{ij}\) as above, DBSCAN yields correct clusters for \(\varepsilon \in [0.01, 0.42]\).
By setting \(k= 2\), the smallest possible value due to the local connectivity constraint, we can further distill the cluster structure down to its bare essentials:
Based on this graph, DBSCAN yields correct clusters for \(\varepsilon \in [0.01, 0.99]\)! Thus, by setting the nearest neighbor parameter of UMAP to a very small value, the cluster separability is dramatically amplified and DBSCAN’s sensitivity w.r.t. \(\varepsilon\) is significantly reduced.
However, the graph layout optimization step has not even been performed yet. This additional step is crucial, in particular for reducing the parameter sensitivity of clustering methods. This is due to the fact \(d_{ij} = 1 - v_{ij}\) only converts affinities into dissimilarities. Finding a graph layout via the cross-entropy \(C_{UMAP}\) as defined in Eq. 4 instead, not only converts affinities (indirectly) into dissimilarities, but also improves the conversion itself w.r.t. to separability (on top of the separability gained by the graph construction), since the optimization procedure optimizes the graph layout for increased cluster separability. This can be explained as follows:
\(C_{UMAP}\) becomes minimal for \(v_{ij} = w_{ij}\). For \(v_{ij} = 0\), the further away from each other the embedding vectors \(y_i\) and \(y_j\) are placed, the better, since this will drive \(w_{ij}\) towards zero. Considering graphs 9 and 10, we see that \(v_{ij}\) is zero mostly for observations from different clusters. Minimizing \(C_{UMAP}\) thus increases cluster separability in the embedding space by driving objects from different clusters apart. Note that minimizing the cross entropy “can be seen as an approximate bound-optimization (or Majorize-Minimize) algorithm [...] implicitly minimizing intra-class distances and maximizing inter-class distances” (Boudiaf et al. 2020, p. 3). The optimization in the graph embedding step of UMAP thus leads to tighter clusters with more white space in between.
The most relevant additional benefit this graph embedding step provides is the large expansion of well-performing \(\varepsilon\)-ranges for DBSCAN. Since the graph layout optimization uses stochastic gradient descent, the resulting embedding vectors are not deterministic. To account for this randomness, we perform 25 embeddings for each value of \(k\) and compute separate averages of the lower and the upper interval boundaries of the \(\varepsilon\)-ranges yielding optimal cluster performance. On average, the obtained embedding coordinates yield correct clusters for \(k= 6\) with \(\varepsilon \in [0.83, 1.03]\), for \(k= 3\) with \(\varepsilon \in [0.70, 6.76]\), and for \(k= 2\) with \(\varepsilon \in [0.79, 20.94]\). Even the smallest (optimal) \(\varepsilon\)-ranges we observed over the \(3 \times 25\) replications are at least as large as the ones obtained on the fuzzy graph for \(k = 6\), and still considerably larger for \(k = 3\) and \(k = 2\): [0.94, 1.03], [0.72, 1.33], [1.16, 4.57], respectively. Further analysis of the variability resulting from optimizing embedding vectors via SGD can be found in “Appendix B”.
These results indicate how crucial optimizing separability by computing embedding vectors is for clustering performance. “Appendix C” confirms its importance on real data.
In these and the following experiments, all of UMAP’s other hyperparameters were left to the implementation defaults, in particular min_dist = 0.1. Additionally adjusting these parameters might further increase separability. However, tuning parameters in an unsupervised setting is a notoriously difficult task and since the results are already convincing by setting \(k\) to a small value, we concentrate on the effect of \(k\).
Summarizing, both the graph construction and the graph embedding steps in the UMAP algorithm independently contribute to an increased separability of clusters in a dataset, and their combined effect improves clusterability dramatically.
4 The price to pay: structures preserved and lost
As we have outlined in the previous sections, UMAP is able to infer and even enhance the topological, i.e. the cluster, structure of a dataset. However, these improvements come at a price which will be outlined in this section.
4.1 Topology versus geometry
Beyond topological structure, i.e., mere “connectedness”, datasets also have geometrical structure—the shapes of the clusters and how the clusters are positioned relative to each other in the ambient space.
Consider the example of a dataset consisting of three nested spheres embedded in a 3-dimensional (Euclidean) space (see Fig. 2A). What kind of structure does this dataset yield? First of all, from a purely topological perspective, we have three unconnected topological subspaces, i.e. clusters: the three spheres. Moreover, from an additional geometrical perspective, we have information on the shape of the individual clusters: they form spheres, i.e. 2-dimensional surfaces. Finally, we have information on the relative position of the clusters to each other within the ambient feature space: the spheres are nested.
What happens if these data are represented in a 2D UMAP embedding? Since a sphere cannot be isometrically mapped to a 2-dimensional plane, some distortion of the geometric structure will be unavoidable in any 2D embedding. Figure 2B shows that, in fact, most of the geometrical structure is lost in UMAP embeddings: the relative positioning of the clusters diverges from the original data and is not consistent over different embeddings.
Effects of UMAP: preservation of topological versus geometrical structure. A: Three nested spheres in 3D (\(n_{{\text {obs}}}= 30000\), part of the data omitted to make the nested structure visible). B: 2D UMAP embeddings for \(k= 7\) and \(k= 15\). The clusters, i.e., topological structure, is preserved. Geometrical structure is not preserved: Ambient space geometry (“nestedness”) is lost; for \(k= 7\), less of the spherical/circular shape is preserved. C: Clustering performances for \(\varepsilon \in [0, 10]\) (step size: 0.01, \(minPts = 5\)) for DBSCAN directly applied to the data (left) and applied to the UMAP embeddings (\(k= 7\): middle, \(k= 15\): right)
The effect on the shape of the clusters is less severe. While for \(k= 15\) the embeddings are similar to circles, i.e. 2D spheres, for \(k= 7\) the general circular shape is retained, yet less uniformly. In contrast, the topological structure of the different clusters are not only preserved in full, but even exaggerated—clusters are much more separated in the embeddings, which is also reflected once again in much wider \(\varepsilon\)-ranges that yield sensible results (Fig. 2C). DBSCAN alone provides perfect clustering performances only over a much smaller \(\varepsilon\) range than when applied to these UMAP embeddings.
As a further example, we consider the complex 2D synthetic dataset by Jain (2010), “who suggest that it cannot be solved by a clustering algorithm” (Barton et al. 2019, p. 2). This “impossible” data contains seven clusters with complex structure, see Fig. 3A. The clusters have different densities, are in part non-convex, and are not linearly separable. DBSCAN by itself is not able to detect the full cluster structure and choosing \(\varepsilon\) from [0, 15] (step size: 0.01, \(minPts = 5\)) based on an optimal ARI value yields a very different cluster result than choosing \(\varepsilon\) based on the optimal NMI value (see Fig. 3B and C). This challenging example further demonstrates two important points:
First, how successfully UMAP embeddings preserve the connected components (i.e. topological structure) and simultaneously distort geometric structure. In Fig. 3D, we can see that the nested structure of the circles and the entanglement of the spirals are completely lost and that the spirals have been “unrolled” in the embedding space, but the different clusters are very clearly separated.
Second, the example illustrates that “dimension inflation” via UMAP can have a positive effect on cluster performance. “Dimension inflation” means that the data is embedded into a space of higher dimensionality than the observed data. Although this is uncommon and we are not aware of any work where this has been investigated before, there are no restrictions that prevent UMAP from being used in this way. Consider Fig. 3F, which shows ARI- and NMI-curves obtained with DBSCAN applied (1) to the data, (2) a 2D UMAP-5, and (3) a 3D UMAP-5 embedding. Although the 2D UMAP-5 embedding already improves performance and strongly reduces parameter sensitivity, it does not yield a perfect solution. In the 2D embedding (Fig. 3D), the two spirals are very close to each other, with a gap between them that is smaller than the gap appearing within the black cluster.
However, the three dimensional UMAP-5 embedding not only further reduces parameter sensitivity, but also allows for perfect cluster performances. A 3D visualization of this embedding is depicted in Fig. 3E, but note that a static 3D visualization does not make the improved separability visible very well. Figures 3G–I show all pairwise plots of the three embedding dimensions of the 3D UMAP embedding, even though none of these 2D projections reflects the cluster structure well. We recommend to base exploratory analysis on 3D embeddings as they are more likely to yield good results in complex data than 2D embeddings and still allow for very reasonable visualizations with dynamic plotting tools.
Another example of complex synthetic data and the beneficial effect of “dimension inflation”. 1st row: the “impossible” data with color according to true cluster structure. 2nd row: data colored according to DBSCAN cluster results if applied directly to the data (different optimal \(\varepsilon\) values for ARI and NMI). 3rd row: Visualizations of a 2D and 3D UMAP-5 embedding with colors according to true cluster structure. 4th row: \(\varepsilon\)-curves for DBSCAN applied to the data, a 2D UMAP-5, and a 3D UMAP-5 embedding. Last row: 2D visualizations of the 3D UMAP-5 embedding with colors according to true cluster structure. In all settings: DBSCAN computed for \(\varepsilon \in [0.01, 15]\), step size: 0.01; \(minPts = 5\)
4.2 Outliers and noise points
Outliers are another important property of a dataset, but their distinctiveness and relative isolation is unlikely to be preserved in their UMAP embeddings. Consider Fig. 4A and C, which shows two 2D datasets with two clusters and, firstly, with two outliers (in blue, A and C), and, secondly, with additional, uniformly distributed noise points (in grey, C). Corresponding UMAP embeddings for \(k= 15\) are depicted in Fig. 4B and D. Although the cluster structure is preserved, in both cases the outliers are no longer detectable as such (note that no dimension reduction has taken place). Similarly for noise points, which are embedded into proximal clusters and then no longer detectable as noise.
It has recently been shown for functional data that outlyingness can be seen as a metric structure of a dataset (Herrmann and Scheipl 2021). Since UMAP does not preserve metric structure (i.e. distances) but connected components, the loss of the outlier structure is not surprising. Moreover, note that UMAP’s local connectivity constraint, which ensures that each point is at least connected to its nearest neighbor, may render it generally impossible to preserve outlier structure in UMAP embeddings. Applying outlier detection methods in an additional preprocessing step before computing UMAP embeddings may solve this issue.
Effect of UMAP on data with outliers and noise points. First column: 2D datasets with two clusters and two outliers (A) and two outliers and noise points (C). Second column: UMAP embeddings with \(k= 15\) (B and D, respectively). The cluster structure is preserved. Outliers and noise points are forced into the clusters
4.3 Overlapping and diffuse clusters
Clusters with considerable overlap or diffuse boundaries that result in a large likelihood of “bridge” points between nominally distinct clusters are especially challenging for most clustering algorithms.
First of all, consider Fig. 5A, which shows a 2D dataset consisting of two clusters which are connected by a small “bridge” of points (blue). From a purely topological perspective, we have a single connected topological subspace. A 2D UMAP representation, however, breaks the connected components apart, see Fig. 5B and C. Note, that this holds for a small value of \(k= 15\) as well as for a very large value of \(k= 505\). Another issue concerns clusters with substantial overlap, which are often modeled as diffuse components of a Gaussian mixture (Rasmussen 2000). In such cases, UMAP and similar manifold learning methods are unlikely to improve clustering performance. Consider Fig. 5D. It shows a 2D dataset with two clusters following 2-dimensional Gaussian distributions with mean vectors \((0, 2)'\) and \((2, 2)'\) and unit covariance matrix. Note that in both embeddings (Fig. 5E and F) the clusters are not clearly separable, and the less so the larger UMAP’s locality parameter \(k\) is chosen.
For strongly overlapping clusters, it is questionable to even consider such settings as (“pure”) clustering tasks. From a topological perspective, such settings cannot be considered a well-posed clustering problem as there are no separable components in the data. However, in the presence of bridges, it seems reasonable to consider the dataset as consisting of two clusters. Whether overlapping clusters should be merged or considered as separate must surely be answered w.r.t. the specific domain. Practitioners should be aware how UMAP tends to behave in such settings: it typically breaks “bridges” apart and merges highly overlapping clusters.
Effect of UMAP on data with connected components. Upper row: 2D data with two bridged clusters. Lower row: 2D dataset with two strongly overlapping clusters. A and D: data. B, C, E, F: UMAP embeddings with \(k= 15\) and \(k= 505\), respectively. UMAP breaks the bridged components up into two clusters, but does not break up the strongly overlapping components
4.4 Quantitative analysis of further synthetic data
In addition to the qualitative analyses of these toy datasets we investigate further examples quantitatively in this paragraph. The datasets under consideration are those from the Fundamental Clustering Problem Suite (FCPS) (Ultsch 2005). These datasets are constructed such that they reflect specific clustering problems. Table 3 shows key characteristics of these datasets and the problems they present. More details including visualizations can be found in the corresponding papers (Thrun and Ultsch 2020; Ultsch and Lötsch 2020). The datasets are provided as a supplement of Ultsch and Lötsch (2020)Footnote 3.
The results of applying DBSCAN directly to the data and on 2D UMAP embeddings with \(k= 10\) are shown in Table 4. Depicted are the highest achievable ARI and NMI values by approach and dataset as well as the \(\varepsilon\)-range \(\varepsilon _{[ARI > 0]}\) for which ARI is greater than zero.
The results show that DBSCAN alone already yields perfect clustering performance for the datasets Hepta, Lsun, Chainlink, Atom, Target, WingNut, and GolfBall. However, note that UMAP clearly reduces \(\varepsilon\) sensitivity (much wider \(\varepsilon\)-range), i.e., it increases clusterability for Hepta, Lsun, Chainlink, Atom, Target. In contrast, on the datasets Tetra and TwoDiamonds, DBSCAN does not perform perfectly. These datasets represent problems (specified as “almost touching clusters” (Tetra) and “cluster borders defined by density” (TwoDiamonds)) with less clearly separable clusters. Consistent with the examples presented in Sect. 3.1, inferring the topological structure via UMAP not only drastically reduces \(\varepsilon\) sensitivity of DBSCAN, it also improves clustering performance to (almost) perfect results in these examples.
However, inferring the relevant structure is not possible with UMAP in the settings EngyTime and Target and thus it does not improve performance of DBSCAN, it even reduces it. This is consistent with the results of the previous subsections: EngyTime is a setting with clusters that overlap strongly, while the Target data is a setting with six clusters of which four are defined by a few outliers.
In summary, the synthetic examples investigated in this and the previous section show that inferring the topological structure of a dataset can dramatically improve and simplify clustering: improvement in the sense that cluster detection with DBSCAN is considerably more reliable, and simplification in the sense that finding good parameters for DBSCAN becomes significantly less challenging: the suitable \(\varepsilon\)-ranges are typically much wider, they consistently start near zero and ARI/NMI quickly reach their optimum in this range, so that a quick and simple coarse grid search over small values of \(\varepsilon\) is likely to be successful. We emphasize that these conclusions apply for diverse and challenging synthetic data settings that include low- as well as high-dimensional data, data with equal and unequal cluster densities, data with (many) irrelevant features, clusters of arbitrary shape and not linearly separable clusters. In the following, we show that this also holds for several real datasets.
5 Experiments on real-world data
An overview of the real datasets used in this study is given in Table 5. Since some of these datasets have already been used in other studies, we can investigate not only how the clustering performance of DBSCAN is improved if the topological structure of a dataset is inferred beforehand. We can additionally compare our results to those reported for other clustering methods. The set of datasets includes the well known Iris data (Anderson 1935; Fisher 1936), the Wine data (Aeberhard et al. 1994; Forina et al. 1988; Dua and Graff 2017), the Pendigits data (Alimoğlu and Alpaydin 2001; Dua and Graff 2017) as well as the COIL (Nane et al. 1996), MNIST (Lecun et al. 1998) and fashion MNIST (FMNIST) (Xiao et al. 2017) data. Following Mukherjee et al. (2019), we use two different versions of FMNIST: one with the original ten clusters and a version reduced to five clusters which are pooled from the original ten based on their similarity. The results of applying DBSCAN directly to the datasets and to the embeddings obtained with UMAP are depicted in Fig. 6 and Table 6.
Figure 6 shows ARI and NMI as a function of \(\varepsilon\) for the different datasets. Table 6 details the optimum ARI and NMI achieved within the considered \(\varepsilon\)-ranges. We inferred the topological structure of the datasets for three different values of \(k\in \{5, 10, 15\}\). Note that we did not tune UMAP at all and used min_dist \(= 0.1\), n_components \(= 3\) and spectral initialization throughout. Iris and Wine data features were scaled respectively standardized.
In general, the results show that what has been observed for the synthetic examples also holds for real data. For all considered settings, inferring the topological structure of the dataset via UMAP before applying DBSCAN leads to better clustering performances than applying DBSCAN directly, dramatically so for MNIST and FMNIST. Moreover, it reduces \(\varepsilon\) sensitivity of DBSCAN with suitable \(\varepsilon\)-ranges starting close to zero and with high (\(> 0.5\)) ARI and NMI values for large parts of the \(\varepsilon\)-range. For DBSCAN directly applied to (F)MNIST, we additionally scanned the \(\varepsilon\)-range [0, 100] with a step size of 0.1, but performance did not improve over this extended search grid.
We also investigate the effect of optimizing the separability by constructing embedding vectors instead of using the fuzzy edge weights directly for datasets Iris, Wine, COIL, and Pendigits. Clustering using UMAP’s fuzzy graph weights directly performs worse, as expected. On the Iris data, e.g., computing embedding vectors with UMAP-10 leads to optimal ARI/NMI \(= 0.89/0.86\) over an \(\varepsilon\)-range of [0.67, 4.82] in contrast to 0.88/0.84 over [0.6, 0.61] if only the fuzzy graph weights of UMAP-10 are used. Both variants still yield better results than applying DBSCAN directly to the data (optimal ARI/NMI \(= 0.75/0.67)\). We found similar results for Wine, COIL, and Pendigits, see “Appendix C”.
In addition, our results show that the fast, simple and very easily tuneable approach we have proposed leads to comparable or superior clustering performances than recently proposed clustering methods such as ClusterGAN (Mukherjee et al. 2019) and SPECTACL(N) (Hess et al. 2019) in some settings. Table 7 lists the highest results obtained on the respective datasets in other studies (Goebl et al. 2014; Mautz et al. 2017; Mukherjee et al. 2019; Hess et al. 2019). On Pendigits and FMNIST-5, DBSCAN applied to UMAP embeddings performs better than the best performing methods FOSSCLU and ClusterGAN as reported by Goebl et al. (2014), Mautz et al. (2017), and Mukherjee et al. (2019). On MNIST, comparable performance is achieved w.r.t. ClusterGAN and better performance w.r.t. SPECTACL(N). Only for the Wine data and FMNIST-10 are better performances reported for methods FOSSCLU, LDA-k-means, and ClusterGAN.
It must be emphasised that these methods also require analysts to pre-specify a fixed number of clusters that are to be found. ClusterGAN’s optimal performances reported in Table 7 were achieved only if the true number of clusters was supplied (Mukherjee et al. 2019). The performance on MNIST considerably deteriorated if the number of clusters was not correctly specified. Recall that one of the major advantages of DBSCAN is that it does not require pre-specifying the number of clusters, in contrast to the complexity of specifying and training ClusterGAN. It should be taken into account, first of all, that a suitable network architecture needs to be defined. Note that standard architectures specified elsewhere had to be adapted for ClusterGAN to achieve satisfactory performance.
In addition, the various hyperparameters for the GAN, the SGD optimizer and the generator-discriminator updating require substantial tuning. Finally, note that our approach works well in settings with both few and many clusters and for both small and large numbers of observations. This is also in contrast to ClusterGAN, which was “particularly difficult [... to train...] with only a few thousand data points” (Mukherjee et al. 2019, p. 4616).
These experiments show that the combination of DBSCAN and UMAP significantly reduces the complexity of finding suitable parameterisations, even in real data. However, the question of how to choose a specific parameterisation requires further discussion, especially since in real exploratory settings there is usually no ground truth to compare the results with.
First of all, it must be emphasised that hyperparameter tuning in unsupervised learning is in general a difficult and unsolved problem, and it is beyond the scope of this study to discuss it in detail. A comprehensive overview of the topic is provided by Zimmermann (2020). As outlined in Sect. 3.1, external evaluation and internal evaluation measures need to be distinguished in cluster analysis. In manifold learning, the performance is often evaluated by comparing the overlap of neighborhoods in the low-dimensional embedding to neighborhoods in the high-dimensional observation space. Overall, however, it remains largely unclear how to properly assess performance and evaluate results (Zimmermann 2020) and much work has been devoted to this question. For clustering see, for example, Ben-David and Ackerman (2008), Vinh et al. (2010b), Rendón et al. (2011), Ullmann et al. (2022) and for manifold learning see, for example, Lee and Verleysen (2008), Lee and Verleysen (2009), Chen and Buja (2009), Rieck and Leitte (2015), Kraemer et al. (2018), Liang et al. (2020). In manifold learning, some work focuses specifically on hyperparameter tuning of certain methods (Alaız et al. 2015; Belkina et al. 2019, e.g.). In contrast, Herrmann and Scheipl (2020) investigate the tunability of different manifold learning methods including UMAP and t-SNE, but with a focus on settings with a single connected manifold.
In the following, we briefly illustrate the complexity of the problem with a simple tuning experiment. Based on the Silhouette Coefficient (Rousseeuw 1987), we performed a grid search to jointly tune UMAP and DBSCAN. Optimizing a performance measure over a grid of parameter settings is a common approach in hyperparameter tuning (Bischl et al. 2023). The Silhouette Coefficient takes into account the mean intra-cluster distance, i.e. the similarity of an object to its cluster (cohesion), and the mean nearest-cluster distance, i.e. the (dis)similarity of an object to other clusters (separation), for each object in a dataset. To evaluate the clustering across all observations we use the Average Silhouette Width (ASW), which takes on values between \(-1\) (worst performance) and 1 (best performance) (Rousseeuw 1987). Since it has been argued above that the use of UMAP in combination with DBSCAN enhances the clusterability in terms of obtaining well-separable clusters, we would expect high ASW for parameterizations that well separate the data into distinct clusters (via UMAP) and that well detect these components (via DBSCAN).
The grid search was performed over different combinations of k and \(\epsilon\). Since the experiments are computationally time demanding, only the datasets Iris, Wine, Pendigits and COIL were used in these experiments. UMAP embeddings were computed for \(k \in \{5, 10, 15,..., 50\}\), which were then clustered with DBSCAN based on 100 different values for \(\epsilon\) equally spaced in [0, 50]. The ASW was then computed for each combination of k and \(\epsilon\). If a clustering resulted in only one cluster, we set the ASW to \(-1\), because it can only be computed for a clustering that yields at least two clusters. Figure 7 shows the results in terms of ASW as a function of \(\epsilon\) grouped by k. The dashed black line reflects the ASW curve obtained for DBSCAN applied directly to the data.
One could now (automatically) select the method parameters without further investigation and visualization by simply using the parameter combination that gives the highest ASW. The advantage of this approach is that it is simple and can be easily extended if other parameters of the two methods are to be included in the optimization. For example, for the COIL data this results in a single optimal parameter configuration of \(k_{opt} = 5\) and \(\epsilon _{opt} = 2.12\), and for Wine with \(k_{opt} = 30\) and \(\epsilon _{opt} = 0.60\).
However, we advise against this approach, firstly because the performance differences between different parameter combinations can be small, and secondly because there may not be a single optimal combination—as is the case for Iris and Pendigtis—as can be seen in Fig. 1. Overall, the results show that high ASW values are obtained over large parts of the \(\epsilon\)-region and that the performance differences between different UMAP embeddings are small in the examples considered. More importantly, while the chosen parameterization may be optimal in terms of ASW, it may not actually be well suited to the problem at hand. In the Pendigits data, the parameter combination \(k = 20\) and \(\epsilon = 41.43\)—the first of several optimal \(\epsilon\)-values – yields an optimal ASW. The corresponding clustering, however, yields only two very unbalanced classes with 10968 and 24 observations, respectively. Given that it is usually assumed that there are 10 clusters, it is very questionable whether this is a desirable parameterization.
In summary, while tuning DBSCAN and UMAP based on grid search and internal evaluation measures may be helpful in practice, we advise against automatic selection of the parameterization. Instead of this naive approach, we would recommend using the results of a grid search to pre-select reasonable parameter configurations, which should then be evaluated in more detail, for example, by inspecting the corresponding embedding visualizations.
6 Discussion
In summary, the presented results show that considering clustering from a topological perspective consistently simplified analysis and improved results in a wide range of settings: from a practical perspective, inferring the topological structure of datasets and representing this structure in suitable embedding vectors that are, in some sense, optimized for separability between the different connected components (dramatically) increased clustering performances of DBSCAN, even outperforming a highly complex deep learning-based clustering method, as long as the clusters did not exhibit large overlap. These insights suggest some conceptual conclusions and raise a number of fundamental questions for cluster analysis, which we will discuss in the following.
To begin with, we argue that two “perspectives” on cluster analysis should be more strictly distinguished: on the one hand, settings were the aim is to infer the number of connected components in a dataset (the “topological perspective”), and, on the other hand, settings where clusters may show considerable overlap (in the following the “probabilistic perspective”). If the “perspective” (implicitly) taken is not clearly specified, the results of a cluster analysis can be misleading. For example, in applied, exploratory analyses relevant information may be lost, while in methodological analyses method comparisons can be misleading.
Consider a truly unsupervised and exploratory setting (i.e. the true number of clusters is not known and determining it is a crucial part of the problem) in an applied context. From the “topological perspective” applying methods that yield a fixed, pre-specified number of clusters is highly questionable in this situation. If the number of clusters is determined a-priori for example via domain knowledge, the analysis cannot falsify these a-priori assumptions about the data and may hide any unexpected structure. This seems contradictory to the purpose of an exploratory analysis, where discovery of unexpected structures can yield valuable new insights. If, on the other hand, approaches such as elbow-plots of cluster quality metrics are used to determine the number of clusters \(n_c\) in a data-driven way, methods inferring and enhancing connected components should be used in the first place.
Another issue concerns the evaluation of competing methods for clustering using datasets with label information. Label information can be misleading, in particular if it is (also) used to pre-specify \(n_c\), as the label information may not be consistent with the unconnected components of a dataset. Consider the FMNIST example, where a simple modification of label information—merging the original 10 into 5 broader categories— leads to considerably different results. Note that this change of labels was not introduced here, but in Mukherjee et al. (2019). Since it requires no specialized domain knowledge to assess the general similarity of clusters in this dataset containing images of pieces of apparel, a change of labels is easy to do. But while this change did not improve the performance of ClusterGAN in terms of ARI and NMI by much, it considerably improves the performance of DBSCAN + UMAP. In other words: the labels were presumably changed such that they were much more consistent with the actual unconnected components—i.e. clusters—in the data. If only the original ten categories of clothing had been considered here, the method comparison would have been misleading, as the different ability of the methods to identify the (un)connected components of the data would have gone unnoticed. The original label information arguably does not reflect the actual cluster structure of the data. This is likely to be the case in many labeled datasets.
On the other hand, consider settings with overlapping clusters. Taking the topological perspective does not make a lot of sense here, as there are no unconnected components if clusters (strongly) overlap and our investigations showed that it is in general questionable that it is possible to infer such cluster structure with methods which aim to infer connected components. In such settings, one should rather take a “probabilistic perspective” and assume that the data follow a joint multi-modal probability distribution, i.e., a mixture of probability distributions. Note that this usually implies some kind of domain knowledge from which it makes sense to assume such structure. Many prominent clustering methods such as k-means, Gaussian Mixture models, or approaches based on the EM algorithm are based on this perspective. It has to be emphasised that our experiments on several widely used real world benchmark datasets showed that an approach based on the topological perspective, which does not use the true number of clusters as a parameter, can perform comparable or even better than methods which do so.
These considerations raise some important questions. First of all, from a rather practical perspective: Is it fair to compare methods which require \(n_c\) as a parameter with those which do not? How trustworthy is the widely used approach to evaluate clustering methods using labeled data? Is it at all useful to apply non-probabilistic clustering methods on data with assumed strong cluster overlap?
Moreover, from a rather general conceptual perspective: Can there be methods which work optimally both in settings with large cluster overlap and settings of high separability? As Schubert et al (2017, p. 19) state in that regard:
“To get deeper insights into DBSCAN, it would also be necessary to evaluate with respect to utility of the resulting clusters, as our experiments suggest that the datasets used do not yield meaningful clusters. We may thus be benchmarking on the ‘wrong’ datasets (but, of course, an algorithm should perform well on any data).”
This already points to the problem of “wrong” datasets, while on the other hand they state a method should perform well in any setting. In the light of the insights presented here, we would argue that it is very fruitful to investigate the characteristics of settings in which a method or combinations of methods works specifically well or even optimally. As outlined, we consider in particular high cluster overlap in contrast to well separable clusters examples of such settings. The underlying principles are fundamentally different (disconnected domains of the clusters versus connected domains of the clusters) and may require different, maybe even contradictory objectives to be optimized. This is specifically relevant as a dataset may consist of both sorts of (assumed) structure. We think the insights and results presented here support this view.
7 Conclusion
This work considered cluster analysis from a topological perspective. Our results suggest that the crucial issue in clustering is not the nominal dimension of the dataset or whether it contains many irrelevant features, but rather how separable the clusters are in the ambient observation space they are embedded in. Extensive experiments on synthetic and real datasets clearly show that focusing on the topological structure of the data can dramatically improve and simplify cluster analysis both in low- and high-dimensional settings. To demonstrate this principle in practice, we used the manifold learning method UMAP to infer the connected components of the datasets and to create embedding vectors optimized for separability, to which we then applied DBSCAN.
Using synthetic data, we showed that this makes results much more robust to hyperparameters in a diverse set of problems including low-dimensional as well as high-dimensional data, data with equal and unequal cluster densities, data with (many) irrelevant features and clusters of arbitrary, not linearly separable shapes. The parameter sensitivity of DBSCAN was consistently and dramatically reduced, simplifying the search for a suitable \(\varepsilon\)-value. Moreover, the cluster detection performance of DBSCAN was considerably improved compared to applying it directly to the data.
Experiments in real data settings corroborated these insights. In addition, our results showed that the simple approach of combining UMAP and DBSCAN can even outperform complex clustering methods SPECTACL and deep-learning-based ClusterGAN on complex image data such as Fashion MNIST.
All these results were obtained with very little hyperparameter tuning for UMAP. In particular, we always used a small value for the number-of-nearest-neighbors parameter denoted k in this work (in most of our experiments: \(k \in \{5, 10, 15\}\)), markedly reducing the complexity of the parameter choice in density-based clustering. All other parameters where set to the default values. Based on a simple toy example we provided a detailed technical explanation why the choice of a small \(k\) is reasonable for the purpose of clustering.
Finally, we propose a conceptual differentiation of cluster analysis suggested by the topological perspective and the presented results. Specifically, we argue that settings with high cluster overlap in contrast to well separable clusters should be considered as fundamentally different settings which require different kinds of methods for optimal results, a distinction usually not made explicit enough. We also propose that using external label information to evaluate clustering solutions should only be done if these labels actually correspond to the (un)connected components of the data manifold from which observations are sampled. If this is not the case, we would argue that evaluation metrics diverge from what clustering algorithms should properly optimize for—identifying (un)connected components—and results will be misleading.
We think these considerations point out important question to be investigated in future work.
Data and code availability
All code and data to reproduce the results can be found on GitHub: https://github.com/HerrMo/topoclust.
Notes
For two matrices \(M_1\) and \(M_2\) with the same dimension, the Hadamard product is defined as \((M_1 \circ M_2)_{ij} = (M_1)_{ij}(M_2)_{ij}\)
The \(L_p\) distance between two vectors \(x = (x_1,..., x_d)^T\) and \(y = (y_1,..., y_d)^T\) in \({\mathbb {R}}^d\) is defined as \(d_p(x, y) = \left( \sum _{i=1}^d(\vert x_i - y_i \vert )^p\right) ^{\frac{1}{p}}\). For \(p = 2\) one obtains the Euclidean distance.
They can be downloaded via http://www.mdpi.com/2306-5729/5/1/13/s1.
References
Aeberhard S, Coomans D, de Vel O (1994) Comparative analysis of statistical pattern recognition methods in high dimensional settings. Pattern Recognit 27(8):1065–1077. https://doi.org/10.1016/0031-3203(94)90145-7
Aggarwal CC (2014) An introduction to cluster analysis. In: Aggarwal CC, Reddy CK (eds) Data clustering, 1st edn. Chapman and Hall/CRC, Boca Raton, pp 1–28. https://doi.org/10.1201/9781315373515
Aggarwal CC (2015) Data mining: the textbook. Springer, Cham. https://doi.org/10.1007/978-3-319-14142-8
Aggarwal CC, Reddy CK (2014) Data clustering: Algorithms and Applications. Chapman and Hall/CRC, Boca Raton. https://doi.org/10.1201/9781315373515
Alaız CM, Fernández Á, Dorronsoro JR (2015) Diffusion maps parameters selection based on neighbourhood preservation. Comput Intell 6
Alimoğlu F, Alpaydin E (2001) Combining multiple representations for pen-based handwritten digit recognition. Turk J Electr Eng Comp Sci 9(1):1–12
Allaoui M, Kherfi ML, Cheriet A (2020) Considerably improving clustering algorithms using UMAP dimensionality reduction technique: a comparative study. International conference on image and signal processing. Springer, Cham, pp 317–325. https://doi.org/10.1007/978-3-030-51935-3_34
Anderson E (1935) The irises of the gaspé peninsula. Bull Am Iris Soc 59:2–5
Ankerst M, Breunig MM, Kriegel HP et al (1999) OPTICS: ordering points to identify the clustering structure. ACM SIGMOD Rec 28(2):49–60. https://doi.org/10.1145/304181.304187
Arias-Castro E, Lerman G, Zhang T (2017) Spectral clustering based on local PCA. J Mach Learn Res 18(9):1–57
Assent I (2012) Clustering high dimensional data. WIREs Data Min Knowl Discov 2(4):340–350. https://doi.org/10.1002/widm.1062
Barton T, Bruna T, Kordik P (2019) Chameleon 2: an improved graph-based clustering algorithm. ACM Trans Knowl Discov Data. https://doi.org/10.1145/3299876
Belkin M, Niyogi P (2001) Laplacian eigenmaps and spectral techniques for embedding and clustering. In: Dietterich T, Becker S, Ghahramani Z (eds) Advances in neural information processing systems, vol 14. MIT Press, Cambridge https://proceedings.neurips.cc/paper/2001/file/f106b7f99d2cb30c3db1c3cc0fde9ccb-Paper.pdf
Belkina AC, Ciccolella CO, Anno R et al (2019) Automated optimized parameters for t-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nature Commun 10(1):5415
Ben-David S, Ackerman M (2008) Measures of clustering quality: a working set of axioms for clustering. In: Koller D, Schuurmans D, Bengio Y, et al (eds) Advances in neural information processing systems, vol 21. Curran Associates, Inc., https://proceedings.neurips.cc/paper/2008/hash/beed13602b9b0e6ecb5b568ff5058f07-Abstract.html
Bengio Y, Courville A, Vincent P (2013) Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 35(8):1798–1828. https://doi.org/10.1109/TPAMI.2013.50
Beyer K, Goldstein J, Ramakrishnan R, et al (1999) When is “nearest neighbor” meaningful? In: Beeri C, Buneman P (eds) Database Theory - ICDT’99. ICDT 1999. Lecture Notes in Computer Science, vol 1540. Springer, Berlin, pp 217–235, https://doi.org/10.1007/3-540-49257-7_15
Bischl B, Binder M, Lang M et al (2023) Hyperparameter optimization: foundations, algorithms, best practices, and open challenges. Wiley Interdiscip Rev Data Min Knowl Discov 13(2):e1484
Blum A, Hopcroft J, Kannan R (2020) Foundations of data science. Cambridge University Press, Cambridge
Boudiaf M, Rony J, Ziko IM, et al (2020) A unifying mutual information view of metric learning: cross-entropy versus pairwise losses. In: Vedaldi A, Bischof H, Brox T, et al (eds) Computer Vision - ECCV 2020. ECCV 2020. Lecture Notes in Computer Science, vol 12351. Springer, Cham, pp 548–564, https://doi.org/10.1007/978-3-030-58539-6_33
Bubenik P (2015) Statistical topological data analysis using persistence landscapes. J Mach Learn Res 16(3):77–102
Campello RJ, Moulavi D, Sander J (2013) Density-based clustering based on hierarchical density estimates. In: Pei J, Tseng V, Cao L, et al (eds) Advances in knowledge discovery and data mining. PAKDD 2013. Lecture Notes in Computer Science, vol 7819. Springer, Berlin, Heidelberg, pp 160–172, https://doi.org/10.1007/978-3-642-37456-2_14
Cayton L (2005) Algorithms for manifold learning. University of California at San Diego, Tech. rep
Chazal F, Michel B (2021) An introduction to topological data analysis: Fundamental and practical aspects for data scientists. Front Artif Intell. https://doi.org/10.3389/frai.2021.667963
Chen L, Buja A (2009) Local multidimensional scaling for nonlinear dimension reduction, graph drawing, and proximity analysis. J Am Stat Assoc 104(485):209–219. https://doi.org/10.1198/jasa.2009.0111
Cohen-Addad V, Schwiegelshohn C (2017) On the local structure of stable clustering instances. arXiv preprint arXiv:1701.08423
Dalmia A, Sia S (2021) Clustering with UMAP: why and how connectivity matters. arXiv preprint https://arxiv.org/abs/2108.05525
Davies DL, Bouldin DW (1979) A cluster separation measure. IEEE Trans Pattern Anal Mach Intell 2:224–227
Debatty T, Michiardi P, Mees W, et al (2014) Determining the k in k-means with MapReduce. In: EDBT/ICDT 2014 Joint Conference, Athènes, Greece, proceedings of the workshops of the EDBT/ICDT 2014 joint conference, https://hal.archives-ouvertes.fr/hal-01525708
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Series B Stat Methodol 39(1):1–22
Doraiswamy H, Tierny J, Silva PJ et al (2021) TopoMap: a 0-dimensional homology preserving projection of high-dimensional data. IEEE Trans Vis Comput Graph 27(2):561–571. https://doi.org/10.1109/TVCG.2020.3030441
Dua D, Graff C (2017) UCI machine learning repository. http://archive.ics.uci.edu/ml
Ester M, Kriegel HP, Sander J, et al (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. In: Proceedings of the second international conference on knowledge discovery and data mining, pp 226–231
Feldman D, Schmidt M, Sohler C (2020) Turning big data into tiny data: constant-size coresets for k-means, PCA, and projective clustering. SIAM J Comput 49(3):601–657. https://doi.org/10.1137/18M1209854
Fisher RA (1936) The use of multiple measurements in taxonomic problems. Ann Eugen 7(2):179–188
Forina M, Leard R, Armanino C, et al (1988) Parvus: an extendible package for data exploration, classification and correlation. Institute of Pharmaceutical and Food Analysis and Technologies, Via Brigata Salerno, 16147 Genoa, Italy
Giordani P, Ferraro MB, Martella F (2020) An introduction to clustering with R, 1st edn. Springer, Singapore. https://doi.org/10.1007/978-981-13-0553-5
Goebl S, He X, Plant C, et al (2014) Finding the optimal subspace for clustering. In: 2014 IEEE international conference on data mining, pp 130–139, https://doi.org/10.1109/ICDM.2014.34
Guan S, Loew M (2021) A novel intrinsic measure of data separability. arXiv preprint https://arxiv.org/abs/2109.05180
Hamerly G, Elkan C (2003) Learning the k in k-means. In: Thrun S, Saul L, Schölkopf B (eds) Advances in neural information processing systems, vol 16. MIT Press, https://proceedings.neurips.cc/paper/2003/file/234833147b97bb6aed53a8f4f1c7a7d8-Paper.pdf
Hastie T, Tibshirani R, Friedman J (2009) The elements of statistical learning: data mining, inference and prediction, 2nd edn. Springer, New York. https://doi.org/10.1007/978-0-387-84858-7
Hennig C, Meila M, Murtagh F et al (2015) Handbook of cluster analysis, 1st edn. Chapman and Hall/CRC, New York. https://doi.org/10.1201/b19706
Herrmann M, Scheipl F (2020) Unsupervised functional data analysis via nonlinear dimension reduction. arXiv preprint arXiv:2012.11987
Herrmann M, Scheipl F (2021) A geometric perspective on functional outlier detection. Stats 4(4):971–1011. https://doi.org/10.3390/stats4040057
Hess S, Duivesteijn W, Honysz P, et al (2019) The SpectACl of nonconvex clustering: a spectral approach to density-based clustering. In: Proceedings of the AAAI conference on artificial intelligence, pp 3788–3795, https://doi.org/10.1609/aaai.v33i01.33013788
Hopkins B, Skellam JG (1954) A new method for determining the type of distribution of plant individuals. Ann Bot 18(2):213–227
Hozumi Y, Wang R, Yin C et al (2021) UMAP-assisted k-means clustering of large-scale SARS-CoV-2 mutation datasets. Comput Biol Med 131(104):264. https://doi.org/10.1016/j.compbiomed.2021.104264
Hubert L, Arabie P (1985) Comparing partitions. J Classif 2(1):193–218. https://doi.org/10.1007/BF01908075
Jain AK (2010) Data clustering: 50 years beyond k-means. Pattern Recognit Lett 31(8):651–666. https://doi.org/10.1016/j.patrec.2009.09.011
Jain AK, Murty MN, Flynn PJ (1999) Data clustering: a review. ACM Comput Surv 31(3):264–323. https://doi.org/10.1145/331499.331504
Kaya IE, Pehlivanlı AÇ, Sekizkardeş EG et al (2017) PCA based clustering for brain tumor segmentation of T1w MRI images. Comput Methods Programs Biomed 140:19–28. https://doi.org/10.1016/j.cmpb.2016.11.011
Kobak D, Linderman GC (2021) Initialization is critical for preserving global data structure in both t-SNE and UMAP. Nat Biotechnol 39(2):156–157. https://doi.org/10.1038/s41587-020-00809-z
Kraemer G, Reichstein M, Mahecha MD (2018) dimRed and coRanking - Unifying Dimensionality Reduction in R. R J 10(1):342. https://doi.org/10.32614/RJ-2018-039
Kriegel HP, Kröger P, Zimek A (2009) Clustering high-dimensional data: a survey on subspace clustering, pattern-based clustering, and correlation clustering. ACM Trans Knowl Discov Data doi 10(1145/1497577):1497578
Kriegel HP, Kröger P, Sander J et al (2011) Density-based clustering. WIREs Data Min Knowl Discov 1(3):231–240. https://doi.org/10.1002/widm.30
Lecun Y, Bottou L, Bengio Y, et al (1998) Gradient-based learning applied to document recognition. In: Proceedings of the IEEE, pp 2278–2324, https://doi.org/10.1109/5.726791
Lee JA, Verleysen M (2007) Nonlinear dimensionality reduction, 1st edn. Springer, New York. https://doi.org/10.1007/978-0-387-39351-3
Lee JA, Verleysen M (2008) Quality assessment of nonlinear dimensionality reduction based on K-ary neighborhoods. In: Saeys Y, Liu H, Inza I, et al (eds) Proceedings of the workshop on new challenges for feature selection in data mining and knowledge discovery at ECML/PKDD 2008, proceedings of machine learning research, vol 4. PMLR, Antwerp, Belgium, pp 21–35
Lee JA, Verleysen M (2009) Quality assessment of dimensionality reduction: rank-based criteria. Neurocomputing 72(7–9):1431–1443. https://doi.org/10.1016/j.neucom.2008.12.017
Liang J, Chenouri S, Small CG (2020) A new method for performance analysis in nonlinear dimensionality reduction. Stat Anal Data Min ASA Data Sci J 13(1):98–108. https://doi.org/10.1002/sam.11445
Linderman GC, Steinerberger S (2019) Clustering with t-sne, provably. SIAM J Math Data Sci 1(2):313–332. https://doi.org/10.1137/18M1216134
Liu J, Han J (2014) Spectral clustering. In: Aggarwal CC, Reddy CK (eds) Data clustering, 1st edn. Chapman and Hall/CRC, Boca Raton, pp 177–200. https://doi.org/10.1201/9781315373515
Lloyd S (1982) Least squares quantization in PCM. IEEE Trans Inf Theory 28(2):129–137. https://doi.org/10.1109/TIT.1982.1056489
Ma Y, Fu Y (eds) (2012) Manifold learning theory and applications, vol 434, 1st edn. CRC Press, Boca Raton. https://doi.org/10.1201/b11431
Mautz D, Ye W, Plant C, et al (2017) Towards an optimal subspace for k-means. In: Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, pp 365–373, https://doi.org/10.1145/3097983.3097989
McInnes L (2018) Using UMAP for Clustering. https://umap-learn.readthedocs.io/en/latest/clustering.html, [Online; accessed 11-January-2022]
McInnes L, Healy J, Melville J (2018) Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint https://arxiv.org/abs/1802.03426
Mehar AM, Matawie K, Maeder A (2013) Determining an optimal value of k in k-means clustering. In: 2013 IEEE international conference on bioinformatics and biomedicine, pp 51–55, https://doi.org/10.1109/BIBM.2013.6732734
Mittal M, Goyal LM, Hemanth DJ et al (2019) Clustering approaches for high-dimensional databases: a review. WIREs Data Min Knowl Discov 9(3):e1300. https://doi.org/10.1002/widm.1300
Mu Z, Wu Y, Yin H, et al (2020) Study on single-phase ground fault location of distribution network based on MDS and DBSCAN clustering. In: 2020 39th Chinese control conference (CCC), IEEE, pp 6146–6150, https://doi.org/10.23919/CCC50068.2020.9188678
Mukherjee S, Asnani H, Lin E, et al (2019) ClusterGAN: latent space clustering in generative adversarial networks. In: Proceedings of the AAAI conference on artificial intelligence, pp 4610–4617, https://doi.org/10.1609/aaai.v33i01.33014610
Nane S, Nayar S, Murase H (1996) Columbia object image library: COIL-20. Department of Computer Science, Columbia University, New York, Tech. rep
Niyogi P, Smale S, Weinberger S (2011) A topological view of unsupervised learning from noisy data. SIAM J Comput 40(3):646–663. https://doi.org/10.1137/090762932
Pandove D, Goel S, Rani R (2018) Systematic review of clustering high-dimensional and large datasets. ACM Trans Knowl Discov Data 12(2):1–68. https://doi.org/10.1145/3132088
Pealat C, Bouleux G, Cheutet V (2021) Improved time-series clustering with UMAP dimension reduction method. In: 2020 25th international conference on pattern recognition (ICPR), IEEE, pp 5658–5665, https://doi.org/10.1109/ICPR48806.2021.9412261
Pham DT, Dimov SS, Nguyen CD (2005) Selection of k in k-means clustering. Proc Inst Mech Eng Part C J Mech Eng Sci 219(1):103–119. https://doi.org/10.1243/095440605X8298
Putri GH, Read MN, Koprinska I et al (2019) Dimensionality reduction for clustering and cluster tracking of cytometry data. In: Tetko IV, Kůrková V, Karpov P et al (eds) Artificial neural networks and machine learning - ICANN 2019: text and time series. ICANN 2019. Lecture notes in computer science, vol 11730. Springer, Cham, pp 624–640. https://doi.org/10.1007/978-3-030-30490-4_50
Rasmussen C (2000) The infinite gaussian mixture model. In: Solla S, Leen T, Müller K (eds) Advances in neural information processing systems, vol 12. MIT Press, Cambridge
Rendón E, Abundez I, Arizmendi A et al (2011) Internal versus external cluster validation indexes. Int J Comput Commun Control 5(1):27–34
Rieck B, Leitte H (2015) Persistent homology for the evaluation of dimensionality reduction schemes. Comput Graph Forum 34(3):431–440. https://doi.org/10.1111/cgf.12655
Riehl E (2011) A leisurely introduction to simplicial sets. Unpublished expository article available online at http://www.math.harvard.edu/eriehl
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65. https://doi.org/10.1016/0377-0427(87)90125-7
Saul LK, Weinberger KQ, Sha F et al (2006) Spectral methods for dimensionality reduction. In: Chapelle O, Schölkopf B, Zien A (eds) Semi-supervised Learning. MIT Press, Cambridge, Massachusetts, pp 293–308
Saxena A, Prasad M, Gupta A et al (2017) A review of clustering techniques and developments. Neurocomputing 267:664–681. https://doi.org/10.1016/j.neucom.2017.06.053
Schubert E, Sander J, Ester M et al (2017) DBSCAN revisited, revisited: Why and how you should (still) use DBSCAN. ACM Trans Database Syst 42(3):1–21. https://doi.org/10.1145/3068335
Scitovski R, Sabo K, Martínez Álvarez F et al (2021) Cluster analysis and applications, 1st edn. Springer, Cham. https://doi.org/10.1007/978-3-030-74552-3
Souvenir R, Pless R (2005) Manifold clustering. In: Tenth IEEE international conference on computer vision (ICCV’05), vol 1, IEEE, pp 648–653, https://doi.org/10.1109/ICCV.2005.149
Tenenbaum JB, De Silva V, Langford JC (2000) A global geometric framework for nonlinear dimensionality reduction. Science 290(5500):2319–2323. https://doi.org/10.1126/science.290.5500.2319
Thrun MC, Ultsch A (2020) Clustering benchmark datasets exploiting the fundamental clustering problems. Data Brief 30(105):501. https://doi.org/10.1016/j.dib.2020.105501
Ullmann T, Hennig C, Boulesteix AL (2022) Validation of cluster analysis results on validation data: a systematic framework. WIREs Data Min Knowl Discov 12(3):e1444. https://doi.org/10.1002/widm.1444
Ultsch A (2005) Clustering with SOM: U*C. In: Proceedings of the workshop on self-organizing maps, Paris, France, https://doi.org/10.13140/RG.2.1.2394.5446
Ultsch A, Lötsch J (2020) The fundamental clustering and projection suite (FCPS): a dataset collection to test the performance of clustering and data projection algorithms. Data. https://doi.org/10.3390/data5010013
van der Maaten L, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9(86):2579–2605
Van Mechelen I, Boulesteix AL, Dangl R, et al (2018) Benchmarking in cluster analysis: a white paper. arXiv:1809.10496 [stat]
Vinh NX, Epps J, Bailey J (2010a) Information theoretic measures for clusterings comparison: Variants, properties, normalization and correction for chance. J Mach Learn Res 11(95):2837–2854
Vinh NX, Epps J, Bailey J (2010b) Information theoretic measures for clusterings comparison: variants, properties, normalization and correction for chance. J Mach Learn Res 11(95):2837–2854
Von Luxburg U (2007) A tutorial on spectral clustering. Stat Comput 17(4):395–416. https://doi.org/10.1007/s11222-007-9033-z
Wang Y, Huang H, Rudin C et al (2021) Understanding how dimension reduction tools work: An empirical approach to deciphering t-SNE, UMAP, TriMap, and PaCMAP for data visualization. J Mach Learn Res 22:1–73
Wasserman L (2018) Topological data analysis. Annu Rev Stat Appl 5(1):501–532. https://doi.org/10.1146/annurev-statistics-031017-100045
Wolf FA, Hamey FK, Plass M et al (2019) PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol 20(59):1–9. https://doi.org/10.1186/s13059-019-1663-x
Xiao H, Rasul K, Vollgraf R (2017) Fashion-MNIST: a novel image dataset for benchmarking machine learning algorithms. arXiv preprint arXiv:1708.07747
Zimek A, Vreeken J (2015) The blind men and the elephant: on meeting the problem of multiple truths in data from clustering and pattern mining perspectives. Mach Learn 98(1):121–155. https://doi.org/10.1007/s10994-013-5334-y
Zimmermann A (2020) Method evaluation, parameterization, and result validation in unsupervised data mining: a critical survey. WIREs Data Min Knowl Discov 10(2):e1330. https://doi.org/10.1002/widm.1330
Zomorodian A, Carlsson G (2005) Computing persistent homology. Discrete Comput Geom 33(2):249–274. https://doi.org/10.1007/s00454-004-1146-y
Funding
Open Access funding enabled and organized by Projekt DEAL. This work has been funded by the German Federal Ministry of Education and Research (BMBF) under Grant No. 01IS18036A. The authors of this work take full responsibility for its content.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Responsible editor: Aristides Gionis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A: Sampling variability
To assess the variability of clustering performance across different samples, we generated 50 datasets for each synthetic setting considered in Sect. 3.1, and computed clusterings via DBSCAN applied directly and to 2D UMAP-5 embeddings. Figure 8 shows box plots of the resulting ARI & NMI distributions.
To compute the clusterings, we used (one of) the \(\epsilon\)-values that correspond to the optimal ARI values depicted in Fig. 1, i.e. \(\epsilon _{opt}\), but also examined the parameter values \(\epsilon _{opt} + 0.2\) and \(\epsilon _{opt} - 0.2\). The optimal \(\epsilon\)-values for the four settings \(E_{100}\), \(E_{1000}\), \(U_3\), and \(U_{1003}\) are—in this order—\(\epsilon _{opt} = \{11.32, 42.64, 0.9, 12.48\}\) for DBSCAN applied directly to the data. For DBSCAN applied to 2D UMAP-5 embeddings, we chose \(\epsilon _{opt} = \{0.24, 0.46, 1.41, 0.28\}\). For \(E_{1000}\), \(U_3\), and \(U_{1003}\), this is the smallest \(\epsilon\)-value that gives the optimal ARI value, but note that there are multiple \(\epsilon\)-values that yield optimal ARI in these settings.
The results underline that applying DBSCAN to UMAP embeddings yields clearly better clustering performance. Overall, one obtains consistently better average performances and much lower variability. In particular, the clustering performances obtained with DBSCAN + UMAP-5 are optimal for the setting \(E_{1000}\), and close to optimal for \(U_{1003}\), for most replications. Furthermore, despite using the smallest possible \(\epsilon _{opt}\), the results are again very robust to changes in \(\epsilon\), both for \(\pm 0.2\) for \(E_{1000}\) and \(U_3\), and for \(+0.2\) in the case of \(U_{1003}\).
Appendix B: Embedding variability
In Sect. 3.2, we showed that although the computation of embedding vectors induces some variability with respect to the meaningful \(\varepsilon\)-range, it also leads to considerably improved separability and is therefore crucial from a clustering perspective. Here we provide additional experiments on this which are based on the synthetic settings from Sect. 3.1 and the three smallest (\(n_{{\text {obs}}}< 10^4\) observations) real datasets Iris, Wine, and COIL. We computed 25 embeddings for each of the datasets (and \(k\) values in the case of the real datasets) and corresponding clusterings on \(\varepsilon\)-grids [0.01, 15] and [0.01, 25], respectively, with a step size of 0.01. For each \(\varepsilon\)-value, the individual minimal, mean, and maximal ARI and NMI values are computed over the 25 replications. Figures 9 and 10 depict the corresponding minimum, mean, and maximum ARI and NMI curves. Note that the curves do not reflect a single embedding, but the worst/mean/best case over all 25 embeddings for each individual \(\varepsilon\)-value. In addition, the maximum ARI and NMI values obtained by applying DBSCAN directly to the data are shown as a black dashed horizontal line and the corresponding \(\varepsilon\)-value as a black dashed vertical line.
In summary, the results again show that optimizing embedding vectors induces some variability with respect to the sensible \(\varepsilon\)-range across different embeddings. However, this variability can be neglected if the main focus is on improving cluster detection. First of all, the variability does not affect the fact that the sensible \(\varepsilon\)-ranges start near zero and quickly reach the optimal value, which is in stark contrast to DBSCAN directly applied to the data (see the black dashed horizontal and vertical lines, and Fig. 1). In addition, in all settings the mean ARI and NMI curves are higher on larger parts of the \(\varepsilon\)-ranges as the maximum ARI and NMI for DBSCAN directly applied to the data. Note that, except for UMAP-5 on Iris and UMAP-15 on COIL, this holds for the minimum curves as well!
Maximum, mean and minimum ARI (left column) and NMI (right column) curves summarized over 25 embeddings of the four synthetic settings \(E_{100}, E_{1000}, U_3, U_{1003}\). Note, the curves do not reflect a single embedding, but the worst/mean/optimal case over all 25 embeddings for each individual \(\varepsilon\)-value. The maximum ARI and NMI values obtained by applying DBSCAN directly to the data are shown as a black dashed horizontal line and the corresponding \(\varepsilon\)-value as a black dashed vertical line. DBSCAN computed for \(\varepsilon \in [0.01, 15]\), step size: 0.01; \(minPts = 5\)
Maximum, mean and minimum ARI (A) and NMI (B) curves summarized over 25 embeddings of the Iris, Wine, and COIL data. Note, the curves do not reflect a single embedding, but the worst/mean/optimal case over all 25 embeddings for each individual \(\varepsilon\)-value. The maximum ARI and NMI values obtained by applying DBSCAN directly to the data are shown as a black dashed horizontal line and the corresponding \(\varepsilon\)-value as a black dashed vertical line. DBSCAN computed for \(\varepsilon \in [0.01, 25]\), step size: 0.01; \(minPts = 5\)
Appendix C: Using just the fuzzy graph weights versus using embedding vectors
Figure 11 shows ARI and NMI as a function of \(\varepsilon\) for four of the real datasets. Cluster results were computed using just the fuzzy graph weights, without additionally computing embedding vectors. Converting the graph weights into dissimilarities via \(d_{ij} = 1 - v_{ij}\), \(i \ne j\), means that the meaningful \(\varepsilon\)-range is restricted to [0, 1]. Moreover, the sensible \(\varepsilon\)-ranges (yielding optimal or high ARI/NMI values) are smaller than those resulting based on additionally optimized embedding vectors.
ARI and NMI as a function of \(\varepsilon\) for four of the real datasets. Results obtained by applying DBSCAN on the fuzzy graph computed by UMAP (converted into a dissimilarity matrix via \(d_{ij} = 1 - v_{ij}\), \(i \ne j\), with \(v_{ij}\) an edge weight). Embedding vectors optimized for separability have not been constructed. DBSCAN computed for \(\varepsilon \in [0.01, 1.5]\), step size: 0.01; \(minPts = 5\)
Here we shortly detail this effect for the Wine, COIL, and Pendigits data based on the UMAP-10 results. The Iris data results are exemplary discussed in Sect. 5.
For the Wine data, only computing the fuzzy graph with UMAP-10 leads to optimal ARI/NMI \(= 0.7/0.61\) for a single \(\varepsilon = 0.5/0.52\). In contrast, additionally computing optimized embedding vectors leads to ARI/NMI = 0.81/0.78 for \(\varepsilon \in [0.64, 0.69]/[1.11, 1.16]\). Unlike the Iris and Wine data, the optimal ARI/NMI value for the Pendigits and COIL data is only achievable for a single \(\varepsilon\)-value. Using embedding vectors is nevertheless beneficial. To see this, consider that on Pendigits an ARI/NMI \(> 0.6\) can be obtained over [0.17, 4.04]/[0.16, 4.13] with embedding vectors. Only using the fuzzy graph would mean that an ARI \(> 0.6\) is not at all achievable and NMI \(> 0.6\) only for \(\varepsilon \in [0.48, 0.56]\). Similar holds for COIL, with ARI/NMI \(> 0.6\) for \(\varepsilon \in [0.25, 0.8]/[0.18, 4.07]\) in contrast to [0.52, 0.68]/[0.45, 0.79].
Again, it needs to be emphasized that only using the fuzzy graph still yields better results than applying DBSCAN directly to the data. For example, applying DBSCAN directly to the Wine data yields optimal ARI/NMI \(= 0.44/0.52\).
In summary, these investigations also show that computing embedding vectors optimized for separability on top of the fuzzy graph not only reduces parameter sensitivity of the clustering method, but can also lead to a better clustering performance due to improved separability.
Appendix D: Real data embedding visualizations
Figure 12 shows 2D UMAP-10 embeddings of the real datasets under investigation. Colors correspond to the class labels. As can be seen, the inferred connected components clearly agree with the labels for the most of the datasets. In FMNIST this holds much better for the 5-label-set. However, note that although 3D embeddings are used in the experiments as they are better suited for cluster detection, they are less well suited for static visualizations (see Sect. 4). That is why we depict UMAP-10 embeddings optimized in two dimensions (i.e. \(d = 2\)) here.
Visualizing 2D UMAP-10 embeddings of the real datasets. Note that an embedding dimension of \(d = 2\) was chosen for the purpose of optimal static visualization, in contrast to \(d = 3\) used for better cluster detection in the quantitative experiments in Sect. 5
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Herrmann, M., Kazempour, D., Scheipl, F. et al. Enhancing cluster analysis via topological manifold learning. Data Min Knowl Disc 38, 840–887 (2024). https://doi.org/10.1007/s10618-023-00980-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10618-023-00980-2