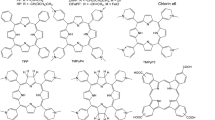
In this work, we analyze the latest data on the molecular docking of a range of SARS-CoV-2 proteins to protoporphyrin IX, verteporfin, and chlorin e6, as well as consider the prospects for using chlorins and porphyrins as agents for photoinactivation of the SARS2 virus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The SARS2 coronavirus became a true scourge in the year 2020. This infectious agent was first identified in China in 2019 and by June 2020 there were practically no countries left unaffected by the pandemic. Apparently, the capabilities of modern medicine remain insufficient for effectively countering the threats arising from novel viruses. The main reasons for the rapid propagation of SARS-CoV-2 include its high contagiousness, the low reliability of rapid tests, and the excessive duration of comprehensive PCR diagnostics, which introduces unacceptable delays in the implementation of quarantine measures. Another reason for the rapid spreading of the COVID-19 is the current lack of drugs capable of completely inactivating coronaviruses in the human body. Coronaviruses are positive-sense RNA viruses belonging to the Coronaviridae family of the order Nidovirales, which are divided into four genera (α, β, γ, and δ). The SARS2 virus belongs to the genus β. Coronaviruses contain four structural proteins: the S-protein (S), the envelope protein (E), the membrane protein (M), and the nucleocapsid protein (N),1 as well as an array of nonstructural (nsp1–16) and accessory proteins (ORF1–10).
There are three strategies in the fight against the SARS2 coronavirus.2 The first strategy involves testing the applicability of existing broad-spectrum antiviral drugs. The advantage of this strategy is that these drugs have been approved for clinical use in humans, thus their metabolic characteristics, potential efficacy, toxicity profile, and the potential side effects have been proved. The disadvantage is that the known antiviral drugs do not directly inactivate the SARS2 coronavirus, and at best can reduce the severity of the disease. The second strategy relies on the search through existing molecular libraries for potential inhibitory or virucidal agents against this coronavirus.3 The third strategy is to develop new, specifically targeted drugs, for example, on the basis of coronavirus genome sequencing. Specifically tailored drugs against the SARS2 virus should clearly be expected to be superior, but the development of a totally new drug molecule from concept to authorization for clinical use can typically take 10–15 years.4
At the present time, most of research efforts follow the second strategy and are devoted to modeling the molecular docking of SARS2 virus proteins with various ligands. The decoded sequences of SARS-CoV-2 proteins enable the application of computer modeling methods to calculate the ligand binding energy of proteins, providing the needed speed of library search for the identification of potential lead compounds for new drug design. There are several well-known molecular docking platforms available, such as SwissDock,5 DockThor,6 AutoDock Vina,7 Surflex,8 GOLD,9 and rDock.10 The objects of modeling are usually the structural and nonstructural proteins of SARS2 virus, however, so far no compound has been found that would be capable of inactivating the SARS2 virus. For example, Cherkasov with coworkers3c reported the binding of more than a billion compounds from the ZINC15 library to the main protease of SARS2 virus.11 The top 1000 ligands – inhibitors of the main protease have been identified, but the list does not contain any porphyrin-like structures. This is rather unexpected, since it is believed that compounds of the porphyrin, chlorin, and phthalocyanine series are among the most promising substances for photoinactivation of viral and drug-resistant bacterial infections.12 The advantages of tetrapyrrole macroheterocyclic compounds over other drugs are that, by binding to a target, they are able to inhibit the virus, and upon subsequent irradiation with light they have virucidal activity. This approach avoids the possibility of mutations or developing drug resistance. Considering the reported strong antiviral activity of protoporphyrin IX and verteporfin against the SARS2 virus,13 the idea of using porphyrins to combat COVID-19 infection is very relevant. An important factor in favor of the use of porphyrins is that a number of compounds belonging to the porphyrin class have been approved for clinical use by the Food and Drug Administration (FDA, USA) and have been also authorized for use in Russia and Europe. When high-affinity binding of porphyrins or their analogs to the proteins of SARS2 virus is achieved, it is possible to develop effective methods for the treatment of coronavirus infections using photoinactivation,14 which is quite convenient to perform in the respiratory tract, and the low dark toxicity of the considered macroheterocyclic compounds is expected to minimize the side effects.
Chlorin e6 belongs to the class of porphyrin compounds and, along with protoporphyrin IX and verteporfin, is already used in clinical practice. Chlorin e6 can be successfully employed in photodynamic therapy, as it generates singlet oxygen with a high quantum yield, absorbs light in the “therapeutic window” and is sufficiently soluble in physiological media.15 Therefore, it can be used for the inactivation of viruses, while the weaker aromaticity of chlorin e6, compared to porphyrins, can provide for stronger afinity toward the proteins of SARS2 virus. Protoporphyrin IX and verteporfin differ by the nature and positions of peripheral substituents, which can also affect the binding localization and energy of tetrapyrrole compound with the proteins of SARS2 virus. This assumption was verified by modeling the binding of chlorin e6, protoporphyrin IX, and verteporfin with a series of SARS-CoV-2 proteins. The following potential targets were selected: the N-terminal RNA-binding domain of nucleocapsid protein (NTD, 6m3m), the S-protein (6vyb), the main protease (6y2e), the ORF3a (6xdc), ORF9b (6z4u), and ORF7A (6w37) proteins, with the selection primarily guided by the biochemical functions of proteins that determine various stages of the virus life cycle. For example, the main functions of the nucleocapsid protein are binding to the viral genome and its packing into a conformation suitable for replication and transcription. During infection, the nucleocapsid protein is prominently expressed, and is capable of eliciting immune response against the SARS2 virus.16 The general domain architecture of the coronavirus nucleocapsid protein consists of three different parts: a highly conserved N-terminal RNAbinding domain (NTD),17 a C-terminal dimerization domain (CTD), and an internally disordered central serine/ arginine linker. The NTD protein of SARS2 virus is a highly promising target for fighting coronavirus infections.
The main protease (Mpro, 3CLpro, nsp5) plays an important role in the translation and replication of new virus generations from viral genomic RNA.18 Many attempts at creating antiviral drugs have targeted viral proteases, but success in this area has been elusive, since the selectivity of such drug candidates was low and the proteases of host cells were also affected, provoking severe complications. The structure of Mpro protease of SARS2 virus is substantially different, while human host cell proteases with such substrate specificity are unknown,19 therefore the Mpro protease of SARS2 virus is a promising target. The S-protein provides binding of angiotensinconverting enzyme 2 of host cell,20 the integration and penetration of virus, as well as the evasion from immune response. The aforementioned proteins are traditional targets and possess the uniqueness required of targets.21
The functions of a large number of additional proteins expressed by open reading frames (ORF) genes for SARS2 virus are still poorly understood or unknown, but the knowledge in this field is rapidly advancing. Thus, for ORF9b, it was found that this protein suppresses innate immunity by acting on the mitochondria and signalosomes MAVS/TRAF3/TRAF6, i.e., suppresses the antiviral transcriptional responses of the host.22
It is known that some of the most severe manifestations of COVID-19 are pneumonia and pronounced anemia,23 therefore it has been hypothesized24 that the virus directly attacks hemoglobin, destroys it, and uses the resulting protoporphyrin to penetrate the host cells. It has been shown that the ORF8 protein and the S-protein of SARS2 virus are capable of binding to porphyrin, while the ORF1ab, ORF10, and ORF3a proteins coordinate the attack on hemoglobin, cause heme demetallization, and then bind the resulting protoporphyrin.24 There is also an alternative opinion, according to which the demetallization of heme and the very process of protein-protein interactions (the interactions of ORF1ab, ORF10, and ORF3a with hemoglobin) are questioned.25 On the other hand, there is experimental evidence that the cleavage of cytochrome с (hemoprotein) is significantly increased in the presence of the ORF3a protein of SARS2 virus.26 On the basis of this, it can be expected that exogenous porphyrin or its analogs can exhibit higher affinity binding to the aforementioned proteins of SARS2 virus, blocking their biological functions and thus inactivating the virus. On the other hand, as it was noted before, the ability of macroheterocyclic compounds to generate reactive oxygen species by the action of light can be used for achieving virucidal effects.27
Accessory proteins encoded by the coronavirus play critical role in the virus-host interactions and modulation of host immune responses, contributing to the pathogenicity of the coronavirus through a variety of strategies. The ORF3a protein acts as a viroporin,28 an ion channel protein, stimulates gene transcription,29 affects the immune response, and induces apoptosis.26,29 It should be noted that the ORF3a protein is highly susceptible to mutations; from this point of view, it is not an optimal target, but it has a sufficient number of conserved domains.26
There is still no consensus view regarding the functions of ORF7a and its role in the life cycle of the virus. It is believed to be involved in protein-protein interactions with host cell proteins and may play role in the viral assembly or budding events30 and evasion of immune response.31 The ORF7a protein has structural homology with protein ICAM-1, which binds to the Т-lymphocyte integrin receptor of LFA-1.32
Thus, the most attractive targets were selected for the study and molecular docking of macroheterocyclic compounds to SARS2 virus proteins was performed to assess their possible inhibitory and virucidal activity. The obtained binding energy values for ligands with the selected proteins are shown in Table 1, while Table 2 shows the most probable hydrogen bonds and π–π interactions between the ligands and amino acid residues of the respective proteins.
The interaction energies of N-terminal RNA-binding domain of the nucleocapsid protein with chlorin e6 and protoporphyrin IX were shown to be rather similar. This was due to the close locations of the aforementioned ligands in the protein. Both compounds formed hydrogen bonds between the peripheral substituents of the macrocycle and the amino acid residues Asn76 and Asn155. In the case of protoporphyrin IX, the energy was slightly higher, because hydrogen bond formation was possible between the nitrogen and hydrogen atoms of the reactive site in porphyrin and the His146 residue. The localization of verteporfin in NTD (Fig. 1), as well as the binding energy with verteporfin was lower, compared to chlorin e6 and protoporphyrin IX, despite the fact that besides hydrogen bonds (Table 2) it can also form π–π interactions with the Tyr110 residue.
On the basis of the obtained data, it can be proposed that all studied macrocyclic compounds can inhibit the functions of NTD, while in the case of chlorin e6 virucidal effects can be achieved via photoirradiation. For the complex of verteporfin with NTD, photoirradiation is unlikely to be effective, since π–π interactions reduce the lifetime of the excited triplet state of porphyrin and, accordingly, the quantum yield of singlet oxygen. In the case of protoporphyrin IX complex with NTD, the hydrogen bonding between the nitrogen and hydrogen atoms of the reactive site in porphyrin and the His146 residue is expected to facilitate the dissipation of light energy and to lower the quantum yield of reactive oxygen species upon photoirradiation. For this reason, the studied porphyrins are unlikely to show virucidal effects. The studied porphyrins and chlorin e6 bind to the S-protein in regions located quite far from the receptor-binding domain (RBD) of the S-protein,33 which is responsible for the binding to ACE2 receptor. Therefore, it is unlikely that the studied macrocyclic compounds could inhibit the binding of S-protein to the ACE2 receptor. Macroheterocyclic compounds are located in the S-protein in such a way that they bind two subunits via hydrogen bonds (Table 2, Fig. 2), while not forming π–π- and H-complexes affecting the aromatic system. Therefore, photoirradiation of protein complexes containing macroheterocyclic compounds can produce a virucidal effect due to cascade oxidation of amino acid residues, cross-linking of polypeptide chains, and irreversible conformational changes in the S-protein.34
Chlorin e6 is located within 4 Å from the Arg567, Ile569, and Asp571 residues of subunit A, the Arg44, His49, Ser50, Lys964, Gln965, Ser967, Ser968, and Asn969 residues of subunit B, and the Leu754, Gln755, Gly757, and Ser758 residues of subunit C. Protoporphyrin IX is located next to the Gln762, Arg765, Ala766, Gly769, Ile770, Val772, Glu773, Leu1012, and Arg1019 residues of subunit A and the Asp950, Asn953, Gln954, Gln1010, Ile1013, Arg1014, and Glu1017 residues of subunit C. Verteporfin is located near the Thr302, Leu303, Lys304, Gln314, Thr315, Asn317, Gln957, Asn960, Thr961, Lys964, and Gln965 residues of subunit B and the Asp737, Thr739, Gly757, Ser758, Thr761, Asn764, Arg765, and Thr768 residues of subunit C in the S-protein.
As noted above, the main protease Mpro plays a key role in the viral replication mechanism,35 therefore its inhibition underlies the antiviral activity of various drugs. For example, it has been proposed36 to use compounds GC376 and GC373, which bind covalently with the Cys145 residue of protease Mpro and inhibit RNA replication in cell culture. Despite the fact that the inhibition of virion replication is an important achievement, as noted above, the use of virucidal drugs leading to the destruction of viral proteins is a more promising approach. Of all the studied macroheterocyclic compounds, chlorin e6 and protoporphyrin IX formed the more stable complexes with viral protease (Fig. 3). However, both macrocycles formed a π–π-complex with the Phe294 residue (Table 2), featuring a coplanar orientation of aromatic systems at a distance of 3.5–3.6 Å, therefore virucidal activity was considered to be unlikely in this case. The ability of verteporfin to absorb light in the “therapeutic window” and the high quantum yield of singlet oxygen during photoirradiation can enhance its virucidal properties.
In the case of the ORF3a protein, modeling showed that both porphyrin and chlorin e6 bind to different parts of the protein globule (Fig. 4). Chlorin e6 and verteporfin interact with the β-sheets of protein, while protoporphyrin IX is located near the α-helix. All studied macrocycles form multiple hydrogen bonds between their peripheral substituents and the amino acid residues of ORF3a protein (Table 2). In addition, it should be noted that chlorin e6 has a higher binding energy with the virus protein, compared to exogenous protoporphyrin IX, giving a reason to expect inhibition of virus while preserving hemoglobin.
The modeling of binding between porphyrins or chlorin e6 and the ORF9b protein showed that their localization in the ORF9b protein was similar (Fig. 5), while the binding energy was somewhat higher in the case of chlorin e6. A common characteristic was the low number of hydrogen bonds and the complete absence of π–π interactions. These features are likely to contribute to the photoinactivation of ORF9b protein.
The studied porphyrins and chlorin e6, judging by the calculated binding energies (Table 1), are expected to have a low inhibitory activity toward the ORF7a protein. The presence of π–π interactions in the case of chlorin e6 with the Phe31 residue in protein and the existence of hydrogen bonds involving the atoms of reactive site in porphyrins and the Tyr5 residue (Table 2, Fig. 6) should cause a decreased quantum yield of reactive oxygen species. It can be concluded that the studied macrocycles cannot be effective toward this target.
The performed study illustrated the ability of the investigated macrocycles to bind structural and accessory proteins of the SARS2 virus. The established ability of macrocycles to form sufficiently strong complexes with a broad range of SARS2 virus proteins can provide virucidal effects at various stages of the virus life cycle. The obtained results, detailing potential specific interactions, allowed to propose that chlorin e6 and protoporphyrin IX should exhibit inhibitory effects against the S-protein, Mpro protease, and NTD, while chlorin e6 is expected to act against the ORF3a protein and verteporfin – against the S-protein. The photooxidative capacity of the studied macrocyclic compounds should be reasonably expected against the following SARS2 virus proteins: protoporphyrin IX – against ORF9b, ORF3a, and the S-protein; chlorin e6 – against the ORF9b, ORF3a, S-protein, and NTD; verteporfin – against the ORF9b and ORF3a proteins, Mpro protease, and S-protein. The studies performed can serve as a basis for a targeted experimental investigation of the inhibitory and photooxidative activity of macrocycles against virus proteins. The most promising agent according to its combination of properties (affinity for virus proteins, effective absorption of light in the “therapeutic window”, high quantum yield of singlet oxygen) is chlorin e6. The clear advantage of chlorin e6 is its ability to produce a dual effect (inhibition and photoinactivation), while having been already approved for clinical use.
Experimental
The following structures of SARS-CoV-2 proteins were used in the work: the N-terminal RNA-binding domain of nucleocapsid protein (NTD, 6m3m), the S-protein (6vyb), main protease Mpro (6y2e), the ORF7a (6w37), ORF3a (6xdc), and ORF9b (6z4u) proteins. The structural information files were downloaded from the Protein Data Bank. The structures of chlorin e6 (PubChem CID: 5479494) and protoporphyrin IX (PubChem CID:4971) were obtained from the PubChem compound database. The structure of verteporfin (PubChem CID: 139032859) was minimized with the ORCA 4.0 program37 within the density functional theory framework using a B3LYP basis set.
The molecular docking of proteins with porphyrins and chlorin e6 was accomplished with the AutoDock Vina software7 and visualized with the PyMOL program. The ligand and protein structure files were prepared with the AutoDock 4.2 program, the grid matrix of the docking area was sized to ensure a complete coverage of the protein molecule. Due to the large size of the grid matrix, the exhaustiveness parameter was raised to 256.38
References
Bosch, B. J.; van der Zee, R.; de Haan, C. A. M.; Rottier, P. J. M. J. Virol. 2003, 77, 8801.
(a) Zumla, A.; Chan, J. F. W.; Azhar, E. I.; Hui, D. S. C.; Yuen, K.-Y. Nat. Rev. Drug Discovery 2016, 15, 327. (b) Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; Zheng, M.; Chen, L.; Li, H. Acta Pharm. Sin. B 2020, 10, 766.
(a) de Wilde, A. H.; Jochmans, D.; Posthuma, C. C.; Zevenhoven-Dobbe, J. C.; van Nieuwkoop, S.; Bestebroer, T. M.; van den Hoogen, B. G.; Neyts, J.; Snijder, E. J. Antimicrob. Agents Chemother. 2014, 58, 4875. (b) Dyall, J.; Coleman, C. M.; Hart, B. J.; Venkataraman, T.; Holbrook, M. R.; Kindrachuk, J.; Johnson, R. F.; Olinger, G. G.; Jahrling, P. B.; Laidlaw, M.; Johansen, L. M.; Lear-Rooney, C. M.; Glass, P. J.; Hensley, L. E.; Frieman, M. B. Antimicrob. Agents Chemother. 2014, 58, 4885. (c) Ton, A.-T.; Gentile, F.; Hsing, M.; Ban, F.; Cherkasov, A. Mol. Inf. 2020, 39, 2000028.
Omrani, A. S.; Saad, M. M.; Baig, K.; Bahloul, A.; Abdul-Matin, M.; Alaidaroos, A. Y.; Almakhlafi, G. A.; Albarrak, M. M.; Memish, Z. A.; Albarrak, A. M. Lancet Infect. Dis. 2014, 14, 1090.
Grosdidier, A.; Zoete, V.; Michielin, O. Nucleic Acids Res. 2011, 39, 270.
Santos, K. B.; Guedes, I. A.; Karl, A. L. M.; Dardenne, L. E. J. Chem. Inf. Model. 2020, 60, 667.
Trott, O.; Olson, A. J. J. Comput. Chem. 2010, 31, 455.
Spitzer, R.; Jain, A. N. J. Comput. Aided Mol. Des. 2012, 26, 687.
Verdonk, M. L.; Cole, J. C.; Hartshorn, M. J.; Murray, C. W.; Taylor, R. D. Proteins 2003, 52, 609.
Ruiz-Carmona, S.; Alvarez-Garcia, D.; Foloppe, N.; Garmendia-Doval, A. B.; Juhos, S.; Schmidtke, P.; Barril, X.; Hubbard, R. E.; Morley, S. D. PLoS Comput. Biol. 2014, 10, e1003571.
Sterling, T.; Irwin, J. J. J. Chem. Inf. Model. 2015, 55, 2324.
Lebedeva, N. Sh.; Gubarev, Yu. A.; Koifman, M. O.; Koifman, O. I. Molecules 2020, 25, 4368.
Gu, C.; Wu, Y.; Guo, H.; Zhu, Y.; Xu, W.; Wang, Y.; Li, Y.; Liu, J.; Yuan, Z.; Zhang, R.; Deng, Q.; Qu, D.; Xie, Y.; Zhou, Y.; Sun, Z.; Cai, X. Sci. Bull. 2020.
Dias, L. D.; Blanco, K. C.; Bagnato, V. S. Photodiagn. Photodyn. Ther. 2020, 31, 101804.
(a) Otvagin, V. F.; Nyuchev, A. V.; Kuzmina, N. S.; Grishin, I. D.; Gavryushin, A. E.; Romanenko, Y. V.; Koifman, O. I.; Belykh, D. V.; Peskova, N. N.; Shilyagina, N. Yu. Eur. J. Med. Chem. 2018, 144, 740. (b) Otvagin, V. F.; Kuzmina, N. S.; Krylova, L. V.; Volovetsky, A. B.; Nyuchev, A. V.; Gavryushin, A. E.; Meshkov, I. N.; Gorbunova, Y. G.; Romanenko, Y. V.; Koifman, O. I.; Balalajeva, I. V.; Fedorov, A. Yu. J. Med. Chem. 2019, 62, 11182. (c) Koifman, O. I.; Luk'yanec, E. A.; Morozova, N. B.; Plotnikova, E. A.; Ponomarev, G. V.; Solov'eva, L. I.; Strahovskaya, M. G.; Yakubovskaya, R. I. RU Patent 2536966C1.
(a) Ahmed, S. F.; Quadeer, A. A.; McKay, M. R. Viruses 2020, 12, 254. (b) Oliveira, S. C.; de Magalhães, M. T. Q.; Homan, E. J. Front. Immunol. 2020, 11, 587615.
Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J.; Ding, C.; Ma, X.; Weng, J.; Gao, Y.; He, H.; Jin, T. Biochem. Biophys. Res. Commun. 2020, 527, 618.
Morse, J. S.; Lalonde, T.; Xu, S.; Liu, W. R. ChemBioChem 2020, 21, 730.
Ullrich, S.; Nitsche, C. Bioorg. Med. Chem. Lett. 2020, 30, 127377.
Habibzadeh, P.; Stoneman, E. K. Int. J. Occup. Environ. Med. 2020, 11, 65.
(a) Gordon, D. E.; Jang, G. M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K. M.; O'Meara, M. J.; Rezelj, V. V.; Guo, J. Z.; Swaney, D. L.; Tummino, T. A.; Huttenhain, R.; Kaake, R. M.; Richards, A. L.; Tutuncuoglu, B.; Foussard, H.; Batra, J.; Haas, K.; Modak, M.; Kim, M.; Haas, P.; Polacco, B. J.; Braberg, H.; Fabius, J. M.; Eckhardt, M.; Soucheray, M.; Bennett, M. J.; Cakir, M.; McGregor, M. J.; Li, Q.; Meyer, B.; Roesch, F.; Vallet, T.; Mac Kain, A.; Miorin, L.; Moreno, E.; Naing, Z. Z. C.; Zhou, Y.; Peng, S.; Shi, Y.; Zhang, Z.; Shen, W.; Kirby, I. T.; Melnyk, J. E.; Chorba, J. S.; Lou, K.; Dai, S. A.; Barrio-Hernandez, I.; Memon, D.; Hernandez-Armenta, C.; Lyu, J.; Mathy, C. J. P.; Perica, T.; Pilla, K. B.; Ganesan, S. J.; Saltzberg, D. J.; Rakesh, R.; Liu, X.; Rosenthal, S. B.; Calviello, L.; Venkataramanan, S.; Liboy-Lugo, J.; Lin, Y.; Huang, X.-P.; Liu, Y.-F.; Wankowicz, S. A.; Bohn, M.; Safari, M.; Ugur, F. S.; Koh, C.; Savar, N. S.; Tran, Q. D.; Shengjuler, D.; Fletcher, S. J.; O'Neal, M. C.; Cai, Y.; Chang, J. C. J.; Broadhurst, D. J.; Klippsten, S.; Sharp, P. P.; Wenzell, N. A.; Kuzuoglu-Ozturk, D.; Wang, H.-Y.; Trenker, R.; Young, J. M.; Cavero, D. A.; Hiatt, J.; Roth, T. L.; Rathore, U.; Subramanian, A.; Noack, J.; Hubert, M.; Stroud, R. M.; Frankel, A. D.; Rosenberg, O. S.; Verba, K. A.; Agard, D. A.; Ott, M.; Emerman, M.; Jura, N.; von Zastrow, M.; Verdin, E.; Ashworth, A.; Schwartz, O.; d'Enfert, C.; Mukherjee, S.; Jacobson, M.; Malik, H. S.; Fujimori, D. G.; Ideker, T.; Craik, C. S.; Floor, S. N.; Fraser, J. S.; Gross, J. D.; Sali, A.; Roth, B. L.; Ruggero, D.; Taunton, J.; Kortemme, T.; Beltrao, P.; Vignuzzi, M.; Garcia-Sastre, A.; Shokat, K. M.; Shoichet, B. K.; Krogan, N. J. Nature 2020, 583, 459. (b) Das, S.; Sarmah, S.; Lyndem, S.; Singha Roy, A. J. Biomol. Struct. Dyn. 2020, 1. (c) Yoshimoto, F. K. Protein J. 2020, 39, 198.
(a) Shi, C.-S.; Qi, H.-Y.; Boularan, C.; Huang, N.-N.; Abu- Asab, M.; Shelhamer, J. H.; Kehrl, J. H. J. Immunol. 2014, 193, 3080. (b) Singh, K. K.; Chaubey, G.; Chen, J. Y.; Suravajhala, P. Am. J. Physiol.: Cell Physiol. 2020, 319, C258.
Gattinoni, L.; Chiumello, D.; Caironi, P.; Busana, M.; Romitti, F.; Brazzi, L.; Camporota, L. Intensive Care Med. 2020, 46, 1099.
Liu, W.; Li, H. J. Am. Chem. Soc. 2020, DOI: https://doi.org/10.26434/chemrxiv.11938173.v7.
(a) Read, R. J. 2020, DOI: https://doi.org/10.26434/chemrxiv.11938173. (b) De Martino, A. W.; Rose, J. J.; Amdahl, M. B.; Dent, M. R.; Shah, F. A.; Bain, W.; McVerry, B. J.; Kitsios, G. D.; Tejero, J.; Gladwin, M. T. Haematologica 2020, 105, 2769.
Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.-Y.; Zhou, W.; Qiu, Y.; Zhou, X. Cell. Mol. Immunol. 2020, 17, 881.
(a) Akter, S.; Inai, M.; Saito, S.; Honda, N.; Hazama, H.; Nishikawa, T.; Kaneda, Y.; Awazu, K. Laser Ther. 2019, 28, 245. (b) Zhang, W.; Zhang, A.; Sun, W.; Yue, Y.; Li, H. Medicine 2018, 97, e10864.
Bianchi, M.; Borsetti, A.; Ciccozzi, M.; Pascarella, S. Int. J. Biol. Macromol. 2020, 170, 820.
Issa, E.; Merhi, G.; Panossian, B.; Salloum, T.; Tokajian, S. mSystems 2020, 5, 00266.
Nelson, C. A.; Pekosz, A.; Lee, C. A.; Diamond, M. S.; Fremont, D. H. Structure 2005, 13, 75.
Neches, R. Y.; Kyrpides, N. C.; Ouzounis, C. A. mBio 2021, 12, 03014.
Nizamudeen, Z. A.; Xu, E.-R.; Karthik, V.; Halawa, M.; Arkill, K. P.; Jackson, A. M.; Bates, D. O.; Emsley, J. Biosci. Rep. 2021, 41, BSR20203837.
Liu, Z.; Xiao, X.; Wei, X.; Li, J.; Yang, J.; Tan, H.; Zhu, J.; Zhang, Q.; Wu, J.; Liu, L. J. Med. Virol. 2020, 92, 595.
(a) Lebedeva, N. Sh.; Yurina, E. S.; Gubarev, Yu. A.; Lyubimtsev, A. V.; Syrbu, S. A. J. Photochem. Photobiol., Sect. A: Chem. 2018, 353, 299. (b) Lebedeva, N. Sh.; Yurina, E. S.; Gubarev, Yu. A.; Kiselev, A. N.; Syrbu, S. A. Mendeleev Commun. 2020, 30, 211.
Xue, X.; Yu, H.; Yang, H.; Xue, F.; Wu, Z.; Shen, W.; Li, J.; Zhou, Z.; Ding, Y.; Zhao, Q.; Zhang, X. C.; Liao, M.; Bartlam, M.; Rao, Z. J. Virol. 2008, 82, 2515.
Vuong, W.; Khan, M. B.; Fischer, C.; Arutyunova, E.; Lamer, T.; Shields, J.; Saffran, H. A.; McKay, R. T.; van Belkum, M. J.; Joyce, M. A.; Young, H. S.; Tyrrell, D. L.; Vederas, J. C.; Lemieux, M. J. Nat. Commun. 2020, 11, 4282.
Neese, F. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2018, 8, e1327.
Rentzsch, R.; Renard, B. Y. Briefings Bioinf. 2015, 16, 1045.
This work received financial support from the Russian Foundation for Basic Research (grant 20-04-60108).
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated from Khimiya Geterotsiklicheskikh Soedinenii, 2021, 57(4), 423–431
Rights and permissions
About this article
Cite this article
Koifman, O.I., Lebedeva, N.S., Gubarev, Y.A. et al. Modeling the binding of protoporphyrin IX, verteporfin, and chlorin e6 to SARS-CoV-2 proteins. Chem Heterocycl Comp 57, 423–431 (2021). https://doi.org/10.1007/s10593-021-02920-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10593-021-02920-8