Abstract
Better knowledge of genetic relationships between the Fortymile caribou herd and its neighbors is needed for conservation decision-making in Canada. Here, we contribute the first fine-scale analysis of genetic population structure in nine contiguous caribou herds at the geographic boundaries between Barren-ground and Northern Mountain caribou, and at the Alaska-Yukon border. Using pairwise differentiation metrics, STRUCTURE, and discriminant analysis of principal components (DAPC) to analyze 15 microsatellite loci in 379 caribou, we found complex patterns of genetic differentiation. The Fortymile was the only herd assigned to more than one genetic cluster, indicative of its history as a larger herd whose range expansions and gene flow to other herds were likely important to maintaining diversity across a functioning genetic metapopulation. Some small herds (Chisana, Klaza, and White Mountains) were genetically distinct, while others (Hart River, Clear Creek, Mentasta) exhibited little differentiation from herds they occasionally overlap, including herds assigned to different conservation units (DUs). This genetic connectivity does not result from demographic connectivity, as episodic contact during rut, rather than herd switching, is the likely mechanism. Unusually, one small herd (White Mountains) maintained genetic differentiation despite rut overlap with Fortymile. Our data reveal that some herds with different ecological and behavioral attributes are demographically independent but nonetheless genetically connected. Thus, we suggest that managing caribou for an appropriate level of genetic connectivity, while also supporting herd persistence, will be essential to conserve caribou genetic diversity in the region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Effective conservation of biological diversity below the species level requires biologically-meaningful conservation units and integrated management across jurisdictions. With many caribou (Rangifer tarandus) herds in North America in decline (Festa-Bianchet et al. 2011; Russell et al. 2018), the conservation unit delineated for each herd increasingly determines its legal status and management. Knowledge of genetic relationships is needed to assess “discreteness” of conservation units for caribou (USFWS and NMFS 1996; COSEWIC 2020), and contribute to their delineation alongside ecological, behavioral, geographic, and evolutionary data (Yannic et al. 2016; COSEWIC 2020). Genetic data can also reveal metapopulation structure, which is needed to ensure conservation actions will maintain gene flow and viable population sizes that allow for ongoing evolutionary processes (Weckworth et al. 2013, 2018).
Canada classified caribou Designatable Units (DUs) for conservation in 2011, but lacked the genetic data needed to assess herds in the central Yukon (COSEWIC 2011), including those along the Alaska-Yukon border (hereafter, “AK-YT caribou”). In particular, COSEWIC states that “the south-western boundary of [the Barren-ground DU3] requires resolution with respect to overlap with Northern Mountain populations (DU7), and particularly the assignment of the Forty-Mile population” (COSEWIC 2011). The populations at this DU boundary are the focus of this study.
Three large AK-YT herds near this DU boundary—the Porcupine, Fortymile, and Nelchina (Table 1; Fig. 1)—complete long-distance annual migrations that cross the USA-Canada border. The Porcupine herd is assigned to the Barren-ground DU (hereafter, “BG”), a DU which has dramatically declined and may be listed as Threatened under the federal Species at Risk Act (COSEWIC 2016). The Porcupine herd, however, has increased and is not considered at risk, though it faces threats from climate change and industrial development (Severson et al. 2021). The Fortymile and Nelchina herds have not yet been assigned a DU. Six additional AK-YT herds in the region are orders of magnitude smaller (Table 1). These caribou occupy much smaller annual ranges, exhibit seasonal altitudinal migrations and shorter longitudinal migrations, are larger-bodied, and tend to disperse rather than aggregate during calving. The Hart River, Clear Creek, Klaza, and Chisana herds are designated in the Northern Mountain DU (hereafter, “NM”), which has a status of “Special Concern” (Ray et al. 2015), and the Mentasta and White Mountains herds in Alaska are not assigned a DU. These small herds are likely vulnerable to stochastic events, overharvest, anthropogenic disturbance, and changes in the quantity or quality of core habitats (Environment Canada 2012). In the absence of comprehensive genetic data, the boundary delineated between the BG and NM DUs was based primarily upon ecotypic differences in behavior and habitat use of these herds (COSEWIC 2011).
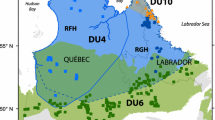
Map of Alaska-Yukon caribou. Polygons represent annual ranges of herds in this study; range overlap does not necessarily indicate co-occurrence because herds may use those portions of their range in different seasons. Colors indicate DU assignment. Cross-hatching represents adjacent herds not included in our study. Dashed line shows a coarse estimate of the former range extent of the Fortymile herd (modified from McDonald and Cooley 2004, Fig. 13); seasonal ranges were much smaller and some portions of the range were infrequently used
Uncertainties about the NM-BG boundary stem from inconsistencies between morphology-based taxonomy (Harding 2022) and phylogenetic, spatial, and ecotypic data. AK-YT caribou descended from a common Beringian-Eurasian lineage (Weckworth et al. 2012; Yannic et al. 2013; Taylor et al. 2021), but divergent evolutionary histories have created genetic substructure within the lineage (Weckworth et al. 2012; Polfus et al. 2017; Taylor et al. 2021) that has not been fully explored. This is especially true for under-studied, small herds that tend to be more genetically complex. Though individual caribou vary in their migratory behavior (Cavedon et al. 2022), and some AK-YT NM individuals demonstrate plasticity in use of seasonal ranges, small NM herds in the AK-YT tend to consistently exhibit shorter, altitudinal migrations and retain fidelity to core portions of their relatively-small ranges even if movement patterns shift in response to changes in habitat or herd density. Inter-herd genetic differentiation is often greater between small herds (Kuhn et al. 2010; Mager et al. 2014; Taylor et al. 2021) than large herds (Cronin et al. 2005; Mager et al. 2014; McFarlane et al. 2016), but because small herds are under-studied, existing data may not accurately represent the fine-scale genetic structure within the BEL. Previous genetic studies of AK-YT caribou either lacked some key herds near the BG-NM boundary (e.g., Kuhn et al. 2010; Taylor et al. 2021) or had small and uneven sample sizes (e.g., Zittlau 2004; Kuhn et al. 2010), which are known to limit the performance of the Bayesian clustering programs used (Puechmaille 2016). Clearly, a fine-scale study of genetic population structure at the boundary of the BG and NM DUs is needed to contribute to assessments of DU discreteness.
The Fortymile and Nelchina herds sit at the nexus of the BG and NM DUs (Fig. 1), but haven’t yet been assigned to a DU. During its once-extensive migrations of 260,000–500,000 caribou in the 1920s (Murie 1935; Boertje et al. 2012), the Fortymile herd overlapped portions of the ranges of most of its neighboring herds (McDonald and Cooley 2004). As the herd declined to ~50,000 in the 1950s-60s and 4,000–6,000 in 1974, its range contracted and shifted (Valkenburg et al. 1994; McDonald and Cooley 2004), which likely reduced gene flow and increased the isolation of some AK-YT herds. Within the past two decades, both the Fortymile and Nelchina herds have expanded their ranges and now utilize large areas of the Yukon, necessitating their assignment to a DU. Genetic data is needed both to inform their DU assignment and to understand how the dramatic changes in abundance and distribution of the Fortymile have influenced gene flow and genetic drift in AK-YT caribou.
In this study, we contribute the first fine-scale analysis of genetic population structure in nine contiguous caribou herds at the boundaries of the barren-ground and northern mountain DUs, and the international border between Alaska and the Yukon. This analysis examines a greater number of microsatellite loci and larger sample sizes per herd than previous studies by combining existing data from 139 caribou (Mager et al. 2014) with 240 new samples from Alaska-Yukon border herds. Using pairwise differentiation metrics and clustering methods, we aim to describe the genetic diversity, differentiation, and population structure of AK-YT herds to contribute to conservation decision-making and interpret patterns of genetic population structure in light of demographic and spatial dynamics over the past century.
Materials and methods
Study area
The study area encompasses approximately 400,000 km2 of Alaska and central Yukon, including subarctic and tundra climate zones. Much of the terrain is mountainous, with boreal forest lowlands and alpine tundra uplands, as well as arctic tundra along the Beaufort Sea coast. Caribou summer ranges tend to be in tundra and subalpine habitats. The size and isolation of these habitat patches influence the spatial distribution of caribou in the region, though habitat selection varies seasonally, annually, and decadally in response to many other factors. Our study focuses on nine contiguous AK-YT herds that span the international border as well as the BG-NM DU boundary (Fig. 1), which are a subset of the approximately 800,000 caribou in 53 herds that inhabit Alaska and the Yukon.
DNA extraction and PCR
Caribou whole blood, dried blood on filter paper, hair, and fecal samples were collected by the Yukon Department of Environment, Alaska Department of Fish and Game, and Bureau of Land Management. The herd identities of sampled caribou were determined by biologists in the natural resource agencies who sampled them, usually based on presence in known seasonal ranges and often confirmed with collar locations. Most samples were from adult females. We extracted DNA from 240 new samples using the DNeasy Blood & Tissue Kit (QIAGEN Inc., Valencia, CA, USA) following manufacturer protocols for each sample type. DNA was amplified at 18 polymorphic microsatellite loci using PCR. Loci were combined in three multiplexes: Multiplex 1—RT6, RT27 and RT1 (Wilson et al. 1997), OheD and OheQ (Jones et al. 2000), NVHRT30 (Røed and Midthjell 1998), BM6506 and BM4513 (Bishop et al. 1994), and OARFCB193 (Buchanan and Crawford 1993); Multiplex 2—RT9long, RT7 and RT24 (Wilson et al. 1997); and Multiplex 3—RT30 (Wilson et al. 1997), BL42 (Bishop et al. 1994), BMS745 (Stone et al. 1995), TEXAN4 (Holder et al. 1994), C89 (Jones et al. 2000) and BMS1788 (Stone et al. 1995). We performed PCR amplifications in 10µL reactions containing 5µL Qiagen Multiplex Master Mix, 2.5µL sterile water, 1µL of the multiplex primer mix and 1.5µL DNA template. Reactions were carried out in an Eppendorf Mastercycler gradient thermocycler or an MJ Research PTC-150 Thermal cycler using the following steps: (1) 5 min initiation at 95 °C followed by (2) 30 cycles of 30 s at 95 °C, 90 s at 57 °C, and 30 s at 72 °C, and (3) a final extension of 45 min at 60 °C. Negative controls were used in each PCR. Fragment lengths were analyzed using a 3730xl 96-Capillary Genetic Analyzer (Applied Biosystems) at the Yale DNA Analysis Facility on Science Hill and the Keck Lab at Yale University (New Haven, CT).
Genotyping
We scored alleles using the Microsat Analysis app (Thermofisher Cloud) with automated bins followed by manual checking of all runs. 92% of samples had duplicate or triplicate genotypes, with 2% of duplicates mismatched on average per locus after low quality samples were excluded. Nearly all inconsistencies were reconciled easily with manual genotyping. When inconsistencies could not be resolved, the data were deleted. In order to calibrate allele calls between these genotypes and existing data from Mager et al. (2014), we re-analyzed 16 archived blood samples. All loci calibrated easily between the two datasets with the exception of BL42, which we eliminated.
We next flagged identical individuals using GenAlEx v.6.503 (Peakall and Smouse 2006, 2012), which is especially important for fecal samples that could be deposited by the same individual (Paetkau 2003). We removed potential duplicate fecal samples including two that were identical, two that were identical at all but 1 locus, and four with missing data that matched other individuals at all remaining loci. A fecal sample from 2018 matched a blood sample collected in 2002 but it is very unlikely that these are the same individual, so both samples were retained.
We checked for systematic genotyping errors including null alleles, allelic dropout, and mis-genotyping of stutter peaks using MICRO-CHECKER v.2.2.3 (Van Oosterhout et al. 2004). The locus RT27 exhibited homozygote excess in three herds, which could be due to allelic dropout or null alleles, so we excluded it. We checked for linkage disequilibrium by testing the null hypothesis of independence of loci in each population and globally using Genepop on the Web v.4.7.5 (Raymond and Rousset 1995; Rousset 2008). We found evidence of linkage between OheD and C89 in a pattern that suggests these two markers may amplify the same locus, so we excluded OheD. After error-checking, the final dataset included genotypes for 379 caribou (Table 1) at 15 microsatellite loci.
Statistical analysis
We used GenoDive v.3.0 (Meirmans and Van Tienderen 2004; Meirmans 2020) to quantify genetic variation by herd, and the pegas package (Paradis 2010) in R v.4.1.0 (R Core Team, 2021) to calculate allelic richness rarefacted to n = 21. We then regressed allelic richness against ln herd size to test for a hypothesized positive relationship (Frankham 1996). We used ADZE v.1.0 (Szpiech et al. 2008) to estimate private allelic richness, rarefacted to the maximum standard sample size, max_G, of n = 21. Private allelic richness was calculated for all possible groupings of herds, in addition to individual herds, with the aim of detecting potential geographic groupings of private alleles.
We used Genodive v.3.0 to calculate metrics of genetic differentiation between pairs of herds, including G’’st (Hedrick 2005), which is standardized to the maximum differentiation possible given the genetic variation at loci, and Jost’s D (Jost et al. 2018), which is independent of within-population diversity. We used 100,000 permutations to compute a p-value for each herd pair, then assessed for significant genetic differentiation after adjusting our alpha of 0.05 for multiple comparisons using two false discovery rate methods (Benjamini and Hochberg 1995; Benjamini and Yekutieli 2001) as well as the Bonferroni correction.
We delineated genetic clusters within our AK-YT caribou sample using the STRUCTURE v.2.3.4 admixture model with correlated allele frequencies and locpriors (Pritchard et al. 2000, 2010; Falush et al. 2003; Hubisz et al. 2009), with herd identity as a proxy for sampling locations because caribou were sampled from known seasonal ranges. The locprior model is well-suited to caribou microsatellites for several reasons: 1) it aids in detecting true clusters when STRUCTURE admixture model results are unclear (Hubisz et al. 2009; Pritchard et al. 2010; Porras-Hurtado et al. 2013); 2) it will not give a false signal of population structure if no structure exists (Hubisz et al. 2009); and 3) it is ideal for datasets with weak population structure that cannot be consistently detected by the standard STRUCTURE model (e.g. populations differentiated by FST < 0.03; Latch et al. 2006). This is particularly important because the maximum value of FST is low in datasets with high within-population variability such as caribou, making it possible for populations that are actually quite differentiated to have low FST, which limits their detection by STRUCTURE. We ran the admixture model without locpriors as well to allow for comparison (Hubisz et al. 2009). All models were run with a burn-in of 200,000 Markov Chain Monte-Carlo (MCMC) iterations, followed by a run length of 500,000 MCMC iterations. The parameter alphapropsd was set at 0.10 to improve mixing after we compared the default (0.025) to alternatives (0.05, 0.10, 0.25, and 0.5) and found that the intermediate values reduced the variability in ln probability across multiple runs at the same K. Because uneven sampling of genetic populations may lead to incorrect clustering results, we also compared an alternative model calculating a separate alpha for each population (Wang 2017) to the default, but results were similar so we used the simpler default model.
We assessed the number of clusters (K) that best represented the underlying population structure using two criteria visualized with Structure Harvester (Earl and vonHoldt 2012): (1) the asymptote of a curve plotting mean log probability against increasing values of K (Pritchard et al. 2010), and (2) ΔK, which is the rate of change of the log probability among values of K (Evanno et al. 2005). Additionally, we visually assessed whether assignment proportions of individuals were asymmetric between populations, and thus likely to represent true population structure (Pritchard et al. 2010). Because STRUCTURE tends to resolve for the highest-order value of K when the ΔK criterion is used (Janes et al. 2017), we used hierarchical clustering to detect potential sub-structure within each cluster (Vähä et al. 2007). After determining the most likely value of K, we examined the individual assignment proportions of the run with the highest log probability (Warnock et al. 2010), grouped all individuals with > 0.50 assignment to each cluster into different datasets, then re-ran each dataset separately. If substructure was detected within a group (i.e., K > 1), the individuals were split again into separate datasets and re-run. For each analysis, we examined models ranging from one to the total number of herds included plus one (e.g., the full sample of nine herds was examined from K = 1–10). Ten replicate runs were performed for each value of K. We used CLUMPAK (Kopelman et al. 2015) to average cluster assignments and align cluster labels across multiple runs for each K.
Because STRUCTURE has known limitations with subtle or complex population structures, it is recommended to compare STRUCTURE results with other approaches (Latch et al. 2006; Janes et al. 2017; Lawson et al. 2018). We implemented discriminant analysis of principal components (DAPC) in the adegenet package v.2.1.3 (Jombart 2008) in R v.4.1.0 (R Core Team, 2021) to summarize patterns of genetic differentiation between herds. DAPC is a multivariate approach that uses PCA to transform genetic data, followed by DA to maximize differentiation between groups, making it useful for clustering subpopulations (Jombart et al. 2010). We used herds as a priori groups rather than assessing the number of groups with the find.clusters() function, which has poor success when differentiation between groups is low (Miller et al. 2020). We retained 90 PCs in order to capture ~ 90% of the variance explained by the PCA. All 8 linear discriminants were retained.
Results
Within-Herd genetic diversity
AK-YT caribou herds were genetically diverse, with expected heterozygosity ranging from 0.79 to 0.85 (Table 2). There was no relationship between ln herd size and allelic richness (simple linear regression: F = 2.10, p = 0.19, R2 = 0.23); some of the smallest herds had low diversity, but other small herds had quite high diversity despite their size. Private allelic richness was generally highest in herds with high total allelic richness (Table 2).
Genetic differentiation
Pairwise genetic differentiation between herds ranged from 0 to 0.18 (Jost’s D), with most herd pairs exhibiting statistically-significant differentiation (Supplementary Information Table S1). The Chisana, Klaza, and White Mountains herds were each strongly and significantly differentiated from all other herds. By contrast, the Hart River and Clear Creek herds were not significantly differentiated from one another or from the neighboring Porcupine or Fortymile herds. The Porcupine and Fortymile herds, however, exhibited low but statistically significant differentiation from each other. Mentasta and Nelchina herds were also significantly differentiated from all other herds (with the exception of Mentasta and Fortymile) but were not differentiated from one another.
Structure analysis
AK-YT caribou were split into two clusters (K = 2) by the STRUCTURE locprior model according to Evanno et al’s (2005) ΔK criterion, with a smaller peak at K = 5 (Supplementary Information Fig. S1), whereas the lowest mean ln probability and its asymptote was at K = 5 (Supplementary Information Fig. S2). The STRUCTURE admixture model without locpriors found K = 1 based on mean ln probability, though an increase at K = 6 could indicate some substructure (Supplementary Information Fig. S3). Similar patterns of genetic clustering were observed in STRUCTURE analyses with and without locpriors, however, the STRUCTURE locprior model more consistently assigned individuals within the same herd to the same clusters at higher proportions of individual assignment (Fig. 2). The values of r < < 1 reported by STRUCTURE for locprior runs also indicated that locations are informative for our data (Hubisz et al. 2009).
Taken together, these STRUCTURE results suggest there are five genetic clusters within our sample, nested within two higher-order genetic clusters (Fig. 2). Hierarchical clustering with the STRUCTURE locprior model found two groups within Cluster 1 and three groups within Cluster 2 based on both the ln probability of the data and ΔK (Fig. 3, Supplementary Information Figs. S4, S5, and S6). Cluster 1 included two subclusters corresponding to the Chisana and Klaza herds (Fig. 3). Within Cluster 2, there was one group comprised of the Clear Creek, Hart River, and Porcupine herds (subcluster 2.1), a second group containing the Mentasta and Nelchina herds (2.2), and a third group containing the White Mountains herd (2.3). All individuals within each of those herds had > 0.50 assignment to their cluster (Fig. 3). The Fortymile herd, however, was not assigned to a single cluster. Most Fortymile individuals had 0.40–0.55 proportion assignment to both subcluster 2.1 and 2.2, and < 0.15 proportion assignment to subcluster 2.3 (Fig. 3). No additional substructure was revealed during a final set of hierarchical STRUCTURE analyses.
Hierarchical clustering of caribou with the STRUCTURE locprior model. Each vertical bar represents an individual, with colors representing the proportion of assignment to each cluster. Step 1: Klaza and Chisana herds cluster separately (Cluster 1) from all other caribou (Cluster 2). Step 2: Cluster 1 caribou were assigned to two subclusters, one containing all Chisana caribou (purple) and the other containing all Klaza caribou (orange). Cluster 2 caribou were assigned to three subclusters: Clear Creek, Hart River, and Porcupine herd individuals all have > 0.50 assignment to subcluster 2.1 (blue); Mentasta and Nelchina herd individuals all have > 0.50 assignment to subcluster 2.2 (burgundy); White Mountains herd individuals all have > 0.50 assignment to subcluster 2.3 (green). Individuals from the Fortymile herd were not assigned to a single cluster; most have 0.40–0.55 assignment to both subcluster 2.1 and 2.2
Discriminant analysis of principal components
In a DAPC, the Chisana herd diverged strongly from other herds. The Klaza and White Mountains herds also diverged from other herds but overlapped one another somewhat when plotted along the first and second discriminant functions (DFs; Fig. 4). Along the third DF, these two herds were also divergent from other herds, but in separate directions (Fig. 4). The Mentasta, Nelchina, and White Mountains herds exhibited partial separation from other herds along the fourth DF (Fig. 4). All other herds overlapped substantially.
Scatterplot showing caribou herds separated along the discriminant functions (DFs) produced by a discriminant analysis of principal components (DAPC). a Herds plotted along the first (x-axis) and second (y-axis) DFs. Symbols represent individual caribou, and ellipses surround two-thirds of the individuals in each herd. Upper-left inset: % of cumulative variance (y-axis) plotted against PCA eigenvalues; shaded area represents 90 eigenvalues retained. Upper-right inset: bar graph of the eight discriminant analysis eigenvalues retained; shaded bars represent the first two discriminant functions the caribou data are plotted on in this figure. b Herds plotted along the third (at left) and fourth (at right) DFs
Discussion
Our analysis of 379 caribou from nine herds revealed complex patterns of genetic population structure in the borderlands of the Yukon and Alaska. Large, migratory caribou herds were, unsurprisingly, the most diverse and least differentiated from other herds in the region. Small caribou herds, however, varied substantially in their genetic diversity and connectivity, even among herds within the same DU, ecotype, and population size. The small Klaza, Chisana, and White Mountains herds were genetically discrete, whereas the small Hart River, Clear Creek, and Mentasta herds could not be genetically differentiated from at least one of their larger neighbors. These patterns were consistent across the STRUCTURE locprior results (Fig. 3), DAPC (Fig. 4), and pairwise herd differentiation (Table S1). All six small herds overlapped or were adjacent to the diverse and historically-large Fortymile herd at times during the past century (McDonald and Cooley 2004), and the extent and seasonality of this contact with Fortymile and other large herds seems to explain the observed diversity and population structure of most herds.
Genetic population structure of Alaska-Yukon herds
The Fortymile herd’s high diversity (Table 2) and partial assignment to multiple genetic clusters (Fig. 3) reflect its past as a much larger herd with an extensive historical range. Its geographic position at the nexus of several mountain ranges with suitable habitat has likely facilitated connectivity with most other herds by allowing for range expansions and shifts during population highs. The genetic structure of AK-YT herds thus reveals a historical genetic metapopulation with the large, migratory Fortymile herd at its center as an agent of genetic connectivity and a repository of genetic diversity.
A severe decline in the Fortymile herd in the past century (Valkenburg et al. 1994; McDonald and Cooley 2004; Boertje et al. 2012), followed by a recovery to less than one-third of its estimated historical peak, has almost certainly altered the dynamics of this genetic metapopulation. A range contraction by the declining Fortymile herd likely reduced gene flow to other herds, and our results could reflect this. For example, STRUCTURE’s failure to assign the Fortymile herd to a single cluster could be an artefact of genetic drift in small and increasingly isolated herds but not in the Fortymile herd. Even if the Fortymile herd lost diversity during its decline, it spent only 5–10 years at − 5,000 individuals before increasing again, which may have limited its loss of allelic diversity (Jangjoo et al. 2016) compared to smaller and increasingly-isolated neighboring herds. However, it is perhaps more likely that our results reflect long term metapopulation structure before Fortymile’s decline. Due to lags in genetic signatures, STRUCTURE results for microsatellites may mirror long-term population dynamics rather than recent population reductions and patterns of gene flow (Weckworth et al. 2012).
Though our results likely reflect dynamics before Fortymile’s recent decline, we found that the genetic diversity and connectivity of AK-YT herds, with the exception of White Mountains herd, could be explained fairly well by their extent, seasonality, and frequency of range overlap with the Fortymile herd over the past century as revealed by local knowledge (McDonald and Cooley 2004) and spatial data from primarily female collared caribou. The small Hart River and Clear Creek herds from the NM DU clustered together with the BG DU Porcupine herd (Fig. 3), consistent with Taylor et al’s (2021) findings from whole-genomes and mtDNA. Both herds also had high within-herd diversity similar to migratory caribou herds (Table 2). Episodic contact during the rut may enable gene flow between these otherwise separate herds. In 2013, 7,000–10,000 Fortymile caribou mixed with Hart River caribou during peak rut in both herds, with tens of thousands of Porcupine caribou arriving later during their own rut, which peaks 3–4 weeks after Hart River/Fortymile (unpublished Government of Yukon data). This likely occurred more frequently in the past (Porcupine Caribou Technical Committee 1993, McDonald and Cooley 2004). Porcupine, Hart River, and Fortymile herds also mix in winter in some years (Caikoski 2020; Lenart 2007). No herd switching from Hart River or Clear Creek into Porcupine or Fortymile has been documented, in contrast to northern Alaska where Porcupine appears to draw some neighboring migratory caribou in Alaska to switch herds. This difference may be due to a preference of migratory caribou from comparatively smaller herds to join large groups (Prichard et al. 2020)—a preference that may not be shared by the less-migratory Hart River and Clear Creek herds. Clear Creek and Hart River caribou, which were once considered a single herd before distinct seasonal ranges were recognized, sometimes overlap and occasionally switch herds (unpublished Government of Yukon data). Some local knowledge suggests Hart River herd was much larger in the past (unpublished Government of Yukon data), which could help to explain its genetic diversity. Taken together, these data suggest that the NM Hart River and Clear Creek herds receive enough gene flow via rut overlap with the Fortymile and BG Porcupine herds to prevent genetic differentiation, but that they remain demographically separate populations.
The two remaining NM DU herds in our study, the Klaza and Chisana herds, were both strongly differentiated from other herds. Ours is the first study to find that Klaza is genetically discrete from Fortymile, likely because we sampled more densely from Klaza than previous studies. Klaza and Chisana had only limited overlap with Fortymile in the past century, and probably not during the rut. The current Klaza summer range was used by Fortymile caribou during winter in the 1920-1930s and 1960s (McDonald and Cooley 2004), and occasionally since 2013. Chisana herd’s current range had even less overlap with the historical Fortymile annual range (McDonald and Cooley 2004), with no clear evidence that the two herds ever co-occurred in the same season, and if so, only in winter. Chisana does occasionally overlap Nelchina and Mentasta herds during the winter (Lieb 1994). We believe a lack of contact with other herds during the rut, coupled with the persistently small size of the Klaza (Hegel 2013) and Chisana herds, likely limits the effective population size (Ne) of each herd enough that genetic drift drives their differentiation from Fortymile. Small Ne can be one of the most important factors explaining spatial genetic structure detected by microsatellites in small, isolated caribou populations (Weckworth et al. 2013). While Chisana and Klaza could have been genetically isolated for long enough that they have developed ecologically-significant local adaptations, it is also possible that the allelic diversity they share with the Fortymile herd (Kuhn et al. 2010) reflects a common genetic history, with recent, rapid genetic drift driving their differentiation.
Three remaining herds in our study, which occur mostly in Alaska and are not assigned to DUs, also give insight into AK-YT metapopulation structure. The small Mentasta and larger migratory Nelchina herds clustered together, consistent with previous microsatellite data, and likely due to rut overlap and male dispersal (Roffler et al. 2012). The Fortymile herd was partially assigned to this Nelchina-Mentasta cluster by STRUCTURE, though we also found low-level differentiation between Nelchina and Fortymile (Table S1). Nelchina and Fortymile ranges overlap during winter (unpublished Government of Yukon data), with increased overlap since the 1990s due to a 100 + km eastward shift by Nelchina from overgrazed former winter range to areas with greater lichen biomass (Collins et al. 2011). Some exchange of individuals, mostly from Nelchina to the Fortymile, has also been observed but does not seem to occur in large numbers (unpublished Government of Yukon data). Interestingly, though Nelchina and Fortymile are large migratory herds currently 100–200 times larger than Mentasta, the Mentasta herd has similar allelic richness and heterozygosity. Mentasta averaged 2,600 individuals in the 1970-80s (Hatcher 2020), whereas Nelchina has signatures of a past genetic bottleneck (Mager et al. 2014), suggesting similar effective population sizes.
Finally, the small White Mountains herd was the only herd we studied that is genetically distinct from Fortymile despite documented periodic overlap in all seasons, including the rut. These findings are surprising given that White Mountains herd was long considered a remnant of the Fortymile, with a shared calving area from 1920 to 1960s (Murie 1935; Skoog 1956; Valkenburg et al. 1994) and rut overlap in the 1960s and since 2008 (Young 2015). Peak rut timing and habitat use are similar for Fortymile and White Mountains, suggesting a different and unknown mechanism separating the herds. If the White Mountains herd has harem-breeding rut behavior (common for small montane herds; Harding 2022) and Fortymile employs short-term pair-bonding behavior common to large migratory herds, genetic differentiation could persist despite spatial overlap during the rut. However, this presumed mating system difference also exists for the Hart River and Porcupine herds that appear to interbreed. It is also possible that frequent rut overlap only began in recent decades. The puzzling differentiation between Fortymile and White Mountains herds highlights a need for further research on how behavioral plasticity and heritable variation in migratory behavior and other traits contribute to ecotypic diversity and taxonomy of Alaskan caribou.
Implications for conservation and assignment of designatable units
Our findings contribute to caribou conservation in Canada by providing the data needed to assess discreteness of conservation units, delineate the boundary between the BG and NM DUs, and assign the Fortymile and Nelchina herds to DUs (COSEWIC 2011). Additionally, our results suggest large herds are drivers of diversity and genetic connectivity in the region, and their capacity for cycles of growth and range expansion that link herds genetically should be considered in conservation decisions.
Our results suggest there is no genetic disjunction at the current boundary between the NM and BG DUs. Instead, the northwestern-most NM herds (Hart River and Clear Creek) grouped with BG caribou, while the NM Klaza and Chisana herds were in separate genetic clusters. Within the scope of our study, there appears to be a genetic break between a northern group of genetically diverse and connected herds (Porcupine, Fortymile, Hart River, and Clear Creek) and herds (e.g., Klaza, Chisana) that do not currently seasonally overlap the ranges of the large migratory herds in our study. COSEWIC (2011) acknowledges that Barren-ground herds tend not to be genetically-discrete, while mountain herds often are. If genetic discreteness among herds is considered a defining feature of the NM DU, then our results could be interpreted as evidence to shift the DU boundary south, with Hart River and Clear Creek becoming part of the BG DU. Genetic data will be needed to evaluate whether adjacent herds such as Bonnet Plume and Redstone could potentially group with the BG DU as well. There are substantial limitations to this idea. First, genetic connectivity between Porcupine and neighboring NM herds does not necessarily indicate demographic connectivity, as episodic contact during the rut is the most likely mechanism preventing genetic divergence of these herds and there is no evidence of significant exchange of individuals between them. Therefore, they may function as a genetic metapopulation but not a demographic one. This distinction is crucial because herds are the management unit used to set sustainable harvest levels that ensure herd persistence and access by local communities to traditional subsistence resources; if herds are demographically independent despite genetic connectivity, then the dynamics within each herd (not inter-herd interactions) shape population trends that are relevant to management. Second, shifting the DU boundary to match genetic population structure ignores important ecological and behavioral differences between the NM and BG herds (Ray et al. 2015), which could potentially reflect heritable variation in migratory behaviors (Cavedon 2022) and ecological traits important for adaptive potential (Polfus et al. 2017). Third, the genetic substructure within the NM DU identified in this study and over a broader area by Taylor et al. (2021) may be evolutionarily-significant in its own right, as it likely reflects a complex evolutionary history including multiple colonization and introgression events during past glacial cycles. It is possible that our focus on the northern portion of the NM DU amplifies the genetic population dynamics at a contact zone between two DUs that are otherwise discrete, though recent work by Taylor et al. (2021) suggests this may not be the case. Fine-scale research throughout the full range of the NM DU would help to clarify these patterns.
Assignment of the Fortymile and perhaps Nelchina herds to a DU is a stated goal of COSEWIC (2011), but we were unable to definitively assign either herd to a DU based on genetics. The Fortymile was the only herd in our study without majority assignment to one of the genetic clusters. The Nelchina herd was somewhat discrete from the Porcupine-Hart River-Clear Creek cluster, but was also quite differentiated from the other herds (except Mentasta, which only rarely ranges into the Yukon and is not assigned a DU). Most individuals in the Fortymile and Nelchina herds undergo long-distance migrations and aggregate for calving, like BG caribou, however both use alpine habitats and have been reduced to less than 10,000 individuals with much smaller ranges in the recent past, similar to NM caribou. We interpret the Fortymile herd’s genetic results as indicative of a once-larger migratory herd that has retained much of its ancestral diversity and signatures of genetic connectivity in the region. The Fortymile may not reach its historical size again soon, however, as the herd has declined since 2017 to ~38,000 caribou. Regardless of what DU the Fortymile herd is assigned to, we suggest that recovery of this large population and its extensive migrations is likely important to sustain a diverse and functioning genetic metapopulation in the AK-YT region.
Conservation of biodiversity includes conservation of evolutionary potential, and our results shed light on how ongoing evolutionary processes operate across DU boundaries in AK-YT caribou. We illuminate three particular ways in which large migratory herds may be important drivers of genetic diversity in small herds in the region:
1) Our findings suggest that occasional contact during the rut has a greater influence on genetic connectivity than annual range overlap in most herds. Importantly, gene flow due to rut overlap occurs without individuals switching herds, thus keeping herds demographically independent. Variation in annual movements by adjacent herds mean that overlap during the brief season of rut only occurs in some years; the genetic structure we observe is likely related to the relative frequency of past rut overlap episodes in most herds (though White Mountains herd is an exception). Thus, seasonality of movements by adjacent herds and their range stability over time may be among the most important factors influencing connectivity between herds. Future shifts in migration routes caused by changes in abundance, environmental change, or human impacts could significantly alter patterns of gene flow in the region.
2) Population size is an influential driver of both genetic diversity and population structure in caribou (Serrouya et al. 2012; Mager et al. 2014). Genetic drift appears to have a strong influence on the genetic discreteness of the smallest AK-YT herds, with the exception of small herds that overlap others during the rut and thus have a larger effective population size than their herd size would suggest. Because of this, infusions of genes from neighboring herds could have an outsized effect on small herds (Frankham et al. 2014).
3) Genetic population structure does not neatly align with ecotype differences in caribou at the NM-BG boundary, but genetics are likely linked with ecotype-associated migratory behaviors via a complex set of feedbacks. Migratory movements and habitat selection differ between individual caribou, with some evidence of a heritable component to these behaviors (Gubili et al. 2017; Cavedon et al. 2022). Nearly all caribou in BG herds and a majority of caribou in NM herds migrate (Cavedon et al. 2022), but NM caribou tend to be more sedentary with shorter migrations and less aggregation. Caribou also sometimes change migratory phenotypes (e.g., caribou translocated from a high-density migratory herd adopted sedentary behavior at lower densities; Hinkes et al. 2005). Sedentary and migratory phenotypes, in turn, shape the geography of herd interactions that affect gene flow. Migratory caribou tend to exhibit greater flexibility in habitat use (Gubili et al. 2017) and can shift their ranges long distances in response to changes in population size (Hinkes et al. 2005) or environmental conditions (Collins et al. 2011).
If large herds drive diversity in the AK-YT region via interactions with neighboring herds, as we propose, it is important to consider how a persistent reduction in the Fortymile herd might impact the genetic diversity, connectivity, and adaptive capacity of other herds in the region. Declines and herd extirpations in threatened and endangered Boreal and Southern Mountain Woodland caribou provide a cautionary tale that suggests viable population sizes and gene flow between herds is crucial. In the Atlantic-Gaspésie caribou population, for example, human activities that reduced gene flow have resulted in two isolated groups of caribou, each with much smaller effective sizes that will likely result in rapid loss of genetic diversity due to drift with increased risk of extirpation (Pelletier et al. 2019). In the Canadian Rockies, where gene flow tends to be restricted to neighboring herds and especially limited for sedentary caribou (Gubili et al. 2017), and where fragmentation of metapopulations limits gene flow (Weckworth et al. 2012), preservation of high-quality habitats that facilitate genetic connectivity is needed. In continuously-distributed Boreal caribou, DU delineation may inadvertently reduce gene flow if management units artificially divide a genetically connected population without ensuring mechanisms for genetic exchange (Priadka et al. 2019). Though NM and BG caribou differ in many ways from boreal herds, similar conservation aims are likely important to AK-YT herds including viable population sizes, ongoing gene flow, and habitat conservation to maintain herd connectivity and enable range expansions, shifts, and behavioral plasticity (Severson et al. 2021). Regardless of how herds are ultimately assigned to DUs, we suggest that supporting the persistence of small herds and conserving intermediary habitats for range expansion of large herds will be crucial to sustain genetic diversity, connectivity, and adaptive potential of caribou in the region.
Data availability
The dataset analyzed in this study is available in the Dryad repository, https://doi.org/10.5061/dryad.gtht76hv1.
Change history
05 June 2024
The sentence "modified from McDonald..." in figure 1 moved to end of the caption.
References
Alaska Department of Fish and Game (2022) New Fortymile caribou population estimate and hunt quota [Press release]. https://www.adfg.alaska.gov/static/applications/webintra/wcnews/2022/releases/08-01-2022.pdf
Alaska Department of Fish and Game (2023) Nelchina caribou hunts RC 561, RC 562, and CC 001 closed by emergency order [Press release]. https://www.adfg.alaska.gov/static/applications/publicnotification/2023/releases/R4-AA-23-1056.pdf
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol) 57:289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Annals Stat 29:1165–1188
Bishop MD, Kappes SM, Keele JW et al (1994) A genetic linkage map for cattle. Genetics 136:619–639. https://doi.org/10.1093/genetics/136.2.619
Boertje RD, Gardner CL, Kellie KA, Taras BD (2012) Fortymile Caribou Herd: increasing numbers, declining nutrition, and expanding range. Alaska Department of Fish and Game, Juneau, Alaska
Buchanan FC, Crawford AM (1993) Ovine microsatellites at the OarFCB11, OarFCB128, OarFCB193, OarFCB266 and OarFCB304 loci. Anim Genet 24:145. https://doi.org/10.1111/j.1365-2052.1993.tb00269.x
Caikoski JR (2020) Porcupine caribou herd management report and plan, Game Management Unit 25A, 25B, 25D, and 26 C: Report period 1 July 2012–30 June 2017, and plan period 1 July 2017–30 June 2022. Alaska Department of Fish and Game, Species Management Report and Plan ADF&G/DWC/SMR&P-2020-22, Juneau
Cavedon M, vonHoldt B, Hebblewhite M et al (2022) Genomic legacy of migration in endangered caribou. PLoS Genet 18(2):e1009974. https://doi.org/10.1371/journal.pgen.1009974
Collins WB, Dale BW, Adams LG et al (2011) Fire, grazing history, lichen abundance, and winter distribution of caribou in Alaska’s taiga. J Wildl Manag 75:369–377. https://doi.org/10.1002/jwmg.39
COSEWIC (2011) Designatable units for Caribou (Rangifer tarandus) in Canada. Committee on the Status of Endangered Wildlife in Canada, Ottawa
COSEWIC (2016) COSEWIC assessment and status report on the Caribou Rangifer tarandus, barren-ground population, in Canada. Committee on the Status of Endangered Wildlife in Canada, Ottawa
COSEWIC (2020) Cosewic Guidelines for Recognizing Designatable Units. https://cosewic.ca/index.php/en-ca/reports/preparing-status-reports/guidelines-recognizing-designatable-units.html. Accessed 6 Jul 2022
Cronin MA, MacNeil MD, Patton JC (2005) Variation in mitochondrial DNA and microsatellite DNA in caribou (Rangifer tarandus) in North America. J Mammal 86:495–505. https://doi.org/10.1644/1545-1542
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Environment Canada (2012) Management plan for the Northern Mountain population of Woodland Caribou (Rangifer tarandus caribou) in Canada. Environment Canada, Ottawa
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. https://doi.org/10.1093/genetics/164.4.1567
Festa-Bianchet M, Ray JC, Boutin S et al (2011) Conservation of caribou (Rangifer tarandus) in Canada: an uncertain future. Can J Zool 89:419–434. https://doi.org/10.1139/z11-025
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508. https://doi.org/10.1046/j.1523-1739.1996.10061500.x
Frankham R, Bradshaw CJA, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63. https://doi.org/10.1016/j.biocon.2013.12.036
Government of Yukon (2018) Yukon state of the environment report. https://open.yukon.ca/sites/default/files/state-environment-report-2018.pdf
Gross JA (2015) Unit 12 caribou. Chapter 7, pages 7 – 1 through 7–11 [In] Harper P, McCarthy LA, editors. Caribou management report of survey and inventory activities 1 July 2012–30 June 2014. Alaska Department of Fish and Game, Species Management ReportADF&G/DWC/SMR-2015-4, Juneau
Gubili C, Mariani S, Weckworth BV et al (2017) Environmental and anthropogenic drivers of connectivity patterns: a basis for prioritizing conservation efforts for threatened populations. Evol Appl 10:199–211. https://doi.org/10.1111/eva.12443
Harding LE (2022) Available names for Rangifer (Mammalia, Artiodactyla, Cervidae) species and subspecies. ZK 1119:117–151. https://doi.org/10.3897/zookeys.1119.80233
Hatcher HL (2020) Mentasta caribou herd management report and plan. Game Management Unit 11: report period 1 July 2012–30 June 2017, and plan period 1 July 2017–30 June 2022. Alaska Department of Fish and Game, Juneau, Alaska, USA
Hatcher HL, Robbins WF (2021) Nelchina caribou herd management report and plan. Alaska Department of Fish and Game, Species Management Report and Plan ADF&G/DWC/SMR&P. -2021-16, JuneauGame Management Unit 13: Report period 1 July 2012–30 June 2017, and plan period 1 July 2017–30 June 2022
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638. https://doi.org/10.1111/j.0014-3820.2005.tb01814.x
Hegel T (2013) Klaza caribou herd inventory studies: 2012 activities. Yukon Fish and Wildlife Branch report PR-13-01. Whitehorse, Yukon, Canada
Hinkes MT, Collins GH, Van Daele LJ et al (2005) Influence of population growth on caribou herd identity, calving ground fidelity, and behavior. J Wildl Manag 69:1147–1162
Holder DA, Arevalo E, Holder MT et al (1994) Bovine microsatellite dinucleotide repeat polymorphisms at the TEXAN-1, TEXAN-2, TEXAN-3, TEXAN-4 and TEXAN-5 loci. Anim Genet 25:201. https://doi.org/10.1111/j.1365-2052.1994.tb00123.x
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332. https://doi.org/10.1111/j.1755-0998.2009.02591.x
Janes JK, Miller JM, Dupuis JR et al (2017) The K = 2 conundrum. Mol Ecol 26:3594–3602. https://doi.org/10.1111/mec.14187
Jangjoo M, Matter SF, Roland J, Keyghobadi N (2016) Connectivity rescues genetic diversity after a demographic bottleneck in a butterfly population network. Proc Natl Acad Sci 113:10914–10919. https://doi.org/10.1073/pnas.1600865113
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Jones KC, Levine KF, Banks JD (2000) DNA-based genetic markers in black-tailed and mule deer for forensic applications. Calif Fish Game 86:115–126
Jost L, Archer F, Flanagan S et al (2018) Differentiation measures for conservation genetics. Evol Appl 11:1139–1148. https://doi.org/10.1111/eva.12590
Kopelman NM, Mayzel J, Jakobsson M et al (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Kuhn TS, Mcfarlane KA, Groves P et al (2010) Modern and ancient DNA reveal recent partial replacement of caribou in the southwest Yukon. Mol Ecol 19:1312–1323. https://doi.org/10.1111/j.1365-294X.2010.04565.x
Latch EK, Dharmarajan G, Glaubitz JC, Rhodes OE (2006) Relative performance of bayesian clustering software for inferringpopulation substructure and individual assignment at low levels of population differentiation. Conserv Genet 7:295–302. https://doi.org/10.1007/s10592-005-9098-1
Lawson DJ, van Dorp L, Falush D (2018) A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat Commun 9:3258. https://doi.org/10.1038/s41467-018-05257-7
Lenart EA (2007) Units 25A, 25B, 25D, and 26 C caribou. Caribou management report of survey and inventory activities 1 July 2004–30 June 2006. Alaska Department of Fish and Game., Juneau, Alaska, USA., pp 232–248
Lieb JW, Cella WB, Tobey RW (1994) Population dynamics of the Mentasta caribou herd. Alaska Department of Fish and Game, Anchorage
Mager KH, Colson KE, Groves P, Hundertmark KJ (2014) Population structure over a broad spatial scale driven by nonanthropogenic factors in a wide-ranging migratory mammal, alaskan caribou. Mol Ecol 23:6045–6057. https://doi.org/10.1111/mec.12999
McDonald J, Cooley D (2004) The historical annual range use patterns of the Fortymile caribou herd. Yukon Fish and Wildlife Branch
McFarlane K, Gunn A, Campbell M et al (2016) Genetic diversity, structure and gene flow of migratory barren-ground caribou (Rangifer tarandus groenlandicus) in Canada. Rangifer 36:1. https://doi.org/10.7557/2.36.1.3577
Meirmans PG (2020) Genodive version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol Ecol Resour 20:1126–1131. https://doi.org/10.1111/1755-0998.13145
Meirmans PG, Van Tienderen PH (2004) Genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794. https://doi.org/10.1111/j.1471-8286.2004.00770.x
Miller JM, Cullingham CI, Peery RM (2020) The influence of a priori grouping on inference of genetic clusters: simulation study and literature review of the DAPC method. Heredity 125:269–280. https://doi.org/10.1038/s41437-020-0348-2
Murie OJ (1935) Alaska-Yukon caribou. U.S. Department of Agriculture, Washington, D.C.
National Park Service (2023) Plans announced for 2023 subsistence hunt of Chisana caribou herd [Press release]. https://www.nps.gov/wrst/learn/news/plans-announced-for-2023-federal-subsistence-hunt-of-chisana-caribou-herd.htm
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Paetkau D (2003) An empirical exploration of data quality in DNA-based population inventories. Mol Ecol 12:1375–1387. https://doi.org/10.1046/j.1365-294X.2003.01820.x
Paradis E (2010) Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420. https://doi.org/10.1093/bioinformatics/btp696
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Pelletier F, Turgeon G, Bourret A et al (2019) Genetic structure and effective size of an endangered population of woodland caribou. Conserv Genet 20:203–213. https://doi.org/10.1007/s10592-018-1124-1
Polfus JL, Manseau M, Klütsch CFC et al (2017) Ancient diversification in glacial refugia leads to intraspecific diversity in a holarctic mammal. J Biogeogr 44:386–396. https://doi.org/10.1111/jbi.12918
Porcupine Caribou Technical Committee (1993) Sensitive habitats of the Porcupine Caribou Herd
Porras-Hurtado L, Ruiz Y, Santos C et al (2013) An overview of STRUCTURE: applications, parameter settings, and supporting software. Front Genet 4:98. https://doi.org/10.3389/fgene.2013.00098
Priadka P, Manseau M, Trottier T et al (2019) Partitioning drivers of spatial genetic variation for a continuously distributed population of boreal caribou: implications for management unit delineation. Ecol Evol 9:141–153. https://doi.org/10.1002/ece3.4682
Prichard AK, Parrett LS, Lenart EA et al (2020) Interchange and overlap among four adjacent Arctic caribou herds. J Wildl Manag 84:1500–1514. https://doi.org/10.1002/jwmg.21934
Pritchard JK, Stephens M, Donnelly P (2000) Inference of Population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Pritchard JK, Wen X, Falush D (2010) Documentation for structure software: Version 2.3
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627. https://doi.org/10.1111/1755-0998.12512
Ray JC, Cichowski DB, St-Laurent M-H et al (2015) Conservation status of caribou in the western mountains of Canada: protections under the species at risk act, 2002–2014. Rangifer 35:49. https://doi.org/10.7557/2.35.2.3647
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. J Hered 86:248–249. https://doi.org/10.1093/oxfordjournals.jhered.a111573
Røed KH, Midthjell L (1998) Microsatellites in reindeer, Rangifer tarandus, and their use in other cervids. Mol Ecol 7:1773–1776. https://doi.org/10.1046/j.1365-294x.1998.00514.x
Roffler GH, Adams LG, Talbot SL et al (2012) Range overlap and individual movements during breeding season influence genetic relationships of caribou herds in south-central Alaska. J Mammal 93:1318–1330. https://doi.org/10.1644/11-MAMM-A-275.1
Rousset F (2008) Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106. https://doi.org/10.1111/j.1471-8286.2007.01931.x
Russell DE, Gunn A, Kutz S (2018) Migratory tundra caribou and wild reindeer. In: Arctic Program. https://arctic.noaa.gov/Report-Card/Report-Card-2018/ArtMID/7878/ArticleID/784/Migratory-Tundra-Rangifer-Caribou-and-Wild-Reindeer. Accessed 5 Jul 2022
Russell KL, Beckmann K, O’Donoghue M, Potié J, Russell KJ (2023) Clear Creek caribou herd population estimate 2018 (SR-23-05). Government of Yukon, Whitehorse, Yukon, Canada
Serrouya R, Paetkau D, McLELLAN BN et al (2012) Population size and major valleys explain microsatellite variation better than taxonomic units for caribou in western Canada. Mol Ecol 21:2588–2601. https://doi.org/10.1111/j.1365-294X.2012.05570.x
Severson JP, Johnson HE, Arthur SM et al (2021) Spring phenology drives range shifts in a migratory Arctic ungulate with key implications for the future. Glob Change Biol 27:4546–4563. https://doi.org/10.1111/gcb.15682
Skoog RO (1956) Range, movements, population, and food habits of the Steese-Fortymile caribou herd. M.Sc. Thesis, University of Alaska Fairbanks
Stone RT, Pulido JC, Duyk GM et al (1995) A small-insert bovine genomic library highly enriched for microsatellite repeat sequences. Mamm Genome 6:714–724. https://doi.org/10.1007/BF00354294
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504. https://doi.org/10.1093/bioinformatics/btn478
Taylor RS, Manseau M, Klütsch CFC et al (2021) Population dynamics of caribou shaped by glacial cycles before the last glacial maximum. Mol Ecol mec 30(23):16166. https://doi.org/10.1111/mec.16166
USFWS, NMFS (1996) Policy regarding the recognition of distinct vertebrate population. segments under the Endangered Species Act
Vähä J-P, Erkinaro J, Niemelä E, Primmer CR (2007) Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol Ecol 16:2638–2654. https://doi.org/10.1111/j.1365-294X.2007.03329.x
Valkenburg P, Kelleyhouse DG, Davis JL, Ver Hoef JM (1994) Case history of the Fortymile Caribou Herd, 1920–1990. Rangifer 14:11. https://doi.org/10.7557/2.14.1.1128
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Wang J (2017) The computer program structure for assigning individuals to populations: easy to use but easier to misuse. Mol Ecol Resour 17:981–990. https://doi.org/10.1111/1755-0998.12650
Warnock WG, Rasmussen JB, Taylor EB (2010) Genetic clustering methods reveal bull trout (Salvelinus confluentus) fine-scale population structure as a spatially nested hierarchy. Conserv Genet 11:1421–1433. https://doi.org/10.1007/s10592-009-9969-y
Weckworth BV, Musiani M, McDevitt AD et al (2012) Reconstruction of caribou evolutionary history in Western North America and its implications for conservation. Mol Ecol 21:3610–3624. https://doi.org/10.1111/j.1365-294X.2012.05621.x
Weckworth BV, Musiani M, DeCesare NJ et al (2013) Preferred habitat and effective population size drive landscape genetic patterns in an endangered species. Proc Royal Soc B: Biol Sci 280:20131756. https://doi.org/10.1098/rspb.2013.1756
Weckworth BV, Hebblewhite M, Mariani S, Musiani M (2018) Lines on a map: conservation units, meta-population dynamics, and recovery of woodland caribou in Canada. Ecosphere 9:e02323. https://doi.org/10.1002/ecs2.2323
Wilson GA, Strobeck C, Wu L, Coffin JW (1997) Characterization of microsatellite loci in caribou Rangifer tarandus, and their use in other artiodactyls. Mol Ecol 6:697–699. https://doi.org/10.1046/j.1365-294x.1997.00237.x
Yannic G, Pellissier L, Ortego J et al (2013) Genetic diversity in caribou linked to past and future climate change. Nat Clim Change 4:132–137. https://doi.org/10.1038/nclimate2074
Yannic G, St-Laurent M-H, Ortego J et al (2016) Integrating ecological and genetic structure to define management units for caribou in Eastern Canada. Conserv Genet 17:437–453. https://doi.org/10.1007/s10592-015-0795-0
Young DD Jr (2015) Unit 20A caribou. Chapter 16. Caribou management report of survey and inventory activities 1 July 2012–30 June 2014. Juneau, Alaska
Zittlau K (2004) Population genetic analyses of North American caribou (Rangifer tarandus). Ph.D. Dissertation, University of Alberta
Acknowledgements
We thank the Yukon Dept of Environment, Alaska Department of Fish and Game, and Bureau of Land Management for providing biological samples. Funding was provided by the Yukon Department of Environment, the Stephen and Sylvia Tregidga Burges Endowed Research Fund and the Stephenson Fund at Earlham College, and Southern Oregon University. Dominic Saidu contributed to laboratory analysis. David Paetkau provided information on genetic markers. Sara Paule, Kim King Jones, Marie Trammell, Daniel Thompson and Nancy Shough supported research logistics and Earlham College and Southern Oregon University provided lab space. We also thank those who contributed to previous research that produced some of our data: Kassidy Colson, Pam Groves, and Kris Hundertmark; North Slope Borough Department of Wildlife Management and U.S. Fish and Wildlife Service for samples; and funding from Alaska EPSCoR NSF award #EPS-0701898 and the State of Alaska, National Fish and Wildlife Foundation award #1499, ADF&G, BLM, and NSF IGERT Resilience and Adaptation Program at University of Alaska Fairbanks.
Funding
This work was funded by the Yukon Department of Environment, the Stephen and Sylvia Tregidga Burges Endowed Research Fund and the Stephenson Fund at Earlham College, and a Southern Oregon University professional development grant.
Author information
Authors and Affiliations
Contributions
K. Mager and M. Suitor conceived of, designed, and obtained funding for the study. M. Suitor, J. Herriges, K. Russell, and J. Stetz secured samples. K. Mager, T. K. Nguyen, and M. Hoang performed lab work and genotyping. K. Mager performed statistical analysis. K. Mager drafted the manuscript, and M. Suitor, J. Herriges, J. Stetz, and K. Russell contributed to the literature review and interpretation of results. All authors revised and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mager, K.H., Suitor, M.J., Nguyen, T.K. et al. Population genetics of caribou in the Alaska-Yukon border region: implications for designation of conservation units and small herd persistence. Conserv Genet 25, 911–924 (2024). https://doi.org/10.1007/s10592-024-01612-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-024-01612-y