Abstract
San Fernando Valley spineflower (Chorizanthe parryi var. fernandina [S. Watson] Jeps.) (Polygonaceae) is an herbaceous annual plant, endemic to California, and until rediscovered in 1999 had been thought to be extinct for almost seven decades. Historically documented at 10 locations, it currently persists at 2, separated by approximately 27 km. State listed as endangered, a description of its genetic diversity and structure is of conservation interest. After determining a lack of variation in ploidy, we examined genetic variation from samples within both populations: a common garden study for potentially adaptive genetic variation in selected growth and phenological traits and analysis of single nucleotide polymorphisms identified through restriction-site associated DNA sequencing. Both measures indicated that this highly restricted taxon nevertheless harbors substantial levels of genetic diversity and has significant between- and within-population genetic structure. Combining approaches from population genomics and common garden studies provided more insight into the patterns and basis of genetic diversity than is typical for studies of non-model species. Although local adaptation was not specifically studied (i.e., via reciprocal transplant studies), the differences determined from these two independent lines of evidence indicate that mixing gene pools between populations is not recommended at this time. Further, with significant differences revealed among subpopulations, we caution against mixing genotypes across subpopulations for the most part, and without much more evidence that this would not pose a risk of outbreeding depression. The importance of supporting pollinator health and diversity is highlighted. With genetic diversity—particularly with an annual species—being dynamic, fluctuating with the usual processes and with contributions from the soil seedbank, we recommend periodic resampling to monitor genetic diversity and structure. Climate change is anticipated to contribute to this variability.
Similar content being viewed by others
Introduction
The effects of rapid climate change manifesting as increased variability in the amount, duration, and seasonal distribution of precipitation and increasing changes in temperature profiles and extremes (Diffenbaugh et al 2015; IPCC 2022), heightens the importance of genetic diversity for providing a means of adjusting to environmental change (Savolainen et al. 2013; Bay et al. 2018; Kelley 2019; Razgour et al. 2019). This may be particularly important for species with limited opportunity for movement in response to environmental change, including small annual plants with limited geographic range. Such restricted species may also consist of few populations, with an increased risk of species extinction as a consequence of extirpation of any of its populations.
One species-rich taxonomic grouping that may be highly dependent for persistence on the availability of appropriate genetic diversity in the face of rapid climate change is Chorizanthe—a New World genus within the buckwheat family (Polygonaceae). Chorizanthe is composed of approximately 10 perennial and 41 annual species, with most annuals found in western North America and mainly in California (Reveal and Hardham 1989). Many of the species are narrowly restricted and seven species (or varieties) are currently state and/or federally listed as threatened or endangered (CNDDB 2022b).
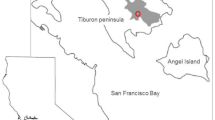
Chorizanthe parryi var. fernandina (S. Watson) Jeps. (San Fernando Valley Spineflower, SFVS, spineflower) is an herbaceous annual, endemic to California and known from ten historical and two current locations (CNPS 2022). After not having been seen since 1929, the taxon was considered extinct for almost seven decades (Hickman 1993; CNPS 2022). SFVS was rediscovered at two locations between 1999 and 2000—at the Ahmanson Ranch in Ventura County (now the Upper Las Virgenes Canyon Open Space Preserve, ULVC) and 27 km (17 mi) north at the Newhall Ranch in Los Angeles County (CNDDB 2022a) (Fig. 1). Following its rediscovery, the U.S. Fish and Wildlife Service (USFWS) designated Chorizanthe parryi var. fernandina (S. Watson) Jeps. as a candidate species for listing (USFWS 1999) and the California Department of Fish and Wildlife (CDFW, formerly California Department of Fish and Game) listed it as endangered under the California Endangered Species Act in 2001 (CDFG 2002). The subsequent inclusion of the Ventura County population as part of the Upper Las Virgenes Canyon Open Space Preserve and mitigation actions that resulted in perpetual protection from development for most of the Newhall population (Dudek 2010) have since reduced habitat conversion as a threat to the taxon.
Although protected from development, SFVS was considered sufficiently vulnerable to extinction to be proposed for listing as threatened under the federal Endangered Species Act (USFWS 2016). Among the ongoing threats described in this rule were competition with non-native invasive plants, climate change, the potential loss of the native pollinator and seed disperser community due to competition with and predation by the non-native Argentine ant (Linepithema humile), vulnerability due to the small number of populations and their disjunct nature, and lack of genetic information. This study was undertaken, in part, to provide information on the genetic status of SFVS, the vulnerability or robustness of the two populations relative to genetic diversity, and recommendations for genetic management. Further, the intent to establish new populations (or subpopulations) using genetic materials from the Newhall Ranch population would benefit from genetic information on the source population.
The two locales that host these two extant populations, ULVC and Newhall Ranch, comprise approximately 18 ha of occupied habitat, although the actual occupied area, as well as numbers of plants, varies annually (USFWS 2004). Within the two known extant populations, SFVS plants occur in patches of several hundred to thousands of plants, typically identified as occurrences (CNDDB 2022a).
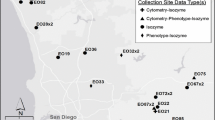
Historic collection records of SFVS include specimens from 10 locations in Los Angeles and Orange County ranging from 15 to 1012 m in elevation (CNDDB 2022a; Fig. 1). Within the extant locales, occurrences are found at elevations 370–430 m (ULVC; CNDDB 2022a) and 292–417 m (Newhall; Dudek 2010). Plants at these locations occur on south-facing slopes at dry, sandy, and sparsely vegetated areas within coastal sage scrub and grasslands, and at the transition zone between these two communities (Fig. 2A–D). Soils at extant sites are associated with a variety of soil units but the locales are similar in that soils are poorly developed and are primarily loams or silty clay loams, and to a lesser extent, sandy loams. Many of these locations have few other native or non-native plants, suggesting that C. parryi var. fernandina (S. Watson) Jeps. is locally adapted to these conditions and might be limited by competition with native and non-native plants elsewhere (Sapphos 2001; Dudek 2010).
There is evidence that SFVS possesses a mixed mating strategy—it is visited by a variety of pollinators and while protandrous, is a facultative self-pollinator (Jones et al. 2009). Flowers are visited by a diversity of potential pollinators, both aerial (e.g., Apis mellifera) and terrestrial (i.e., native ants), which are associated with the surrounding coastal sage scrub community, indicating that SFVS has adopted a generalist pollination strategy (Jones et al. 2009). Seed set is abundant “in the wild” (i.e., in situ and absent manipulations) although both fruit set and seed viability are lower for self-pollinated plants (Jones et al. 2009, 2010). The small seeds, which remain in the involucre after the plant disarticulates, lack morphological modifications to facilitate wind or animal dispersal and are largely dispersed by native ants and small mammals (Sapphos 2001; USFWS 2016). Germination occurs following the onset of seasonal rains and different cohorts emerge from the seed bank though the growing season (Dudek 2010).
Although the life history traits of this taxon have not been thoroughly researched, its characterization as a small stature, annual, endemic plant species with evidence of selfing would suggest that it is likely to exhibit modest within-species genetic diversity and significant population structure (Hamrick and Godt 1989). The geographic distance between the two populations (27 km) is such that gene flow between them seems highly unlikely, expressed as isolation by distance (IBD) (Wright 1943). Any gene flow would be further impeded by the intervening (low) mountainous landscape comprised of the Simi Hills and the Santa Susana Mountains. Apparent or cryptic differences in environmental variables among subpopulations and between populations could have generated genetic differences (isolation by environment [IBE]), including potential local adaptation.
In this study—the first to examine genetic diversity in this taxon—we secured both foliar and seed samples from the two extant populations and employed flow cytometry, restriction site-associated DNA sequencing, and common garden studies to elucidate the nature and structure of genetic diversity. The utility of genetic diversity for providing resilience to rapid climate change may be a function of both the amount and structure of within-species diversity. Further, the nature of genetic diversity—as the extent to which it provides for adaptive response—informs its utility for this purpose. Specifically, our objectives were to: (1) look for evidence of intraspecific ploidy variation, as ploidy levels of known to vary within and between species of the genus Chorizanthe (Hardham 1989); (2) characterize levels of genetic variation and how it is distributed between and within the two extant populations; (3) evaluate the presence of potentially adaptive genetic differences between and within populations; and (4) further characterize the breeding system.
Methods
Study site selection and sample collection
The two extant locations for this spineflower were designated as ‘populations’ for sampling purposes and documentation: Newhall Ranch in Los Angeles County and Upper Las Virgenes Canyon Open Space Preserve (ULVC, Las Virgenes) in Ventura County (Fig. 1, Table 1). Sites were then selected within each population to test for within-population substructure, designating these sites as ‘subpopulations’. ULVC subpopulations were disjunct occurrences located within the full observable extent of that population (approximately 1.6 km); those subpopulations were simply named one through six, from west to east. Distances between adjacent subpopulations ranged from 0.25 to 0.6 km—with the largest distance being between the eastern-most subpopulations (LV5 and LV6). For the Newhall Ranch population, eight subpopulations were selected based on earlier monitoring data that indicated consistent spineflower presence and diversity among sampling sites (e.g., soils, vegetative associates, elevation, and latitude/longitude) (McGraw 2018). During previous monitoring activities, subpopulations were assigned unique names, which we used in this study for consistency. For the Newhall population, distances between adjacent subpopulations used in this study ranged from 0.8 to 3.1 km, across a distance of approximately 8 km for the entire sampling area. Locality information, including latitude and longitude (NAD 83), elevation, and certain soil, slope, vegetative composition, or other defining characteristics are presented for each subpopulation in Table 1.
Prior to sampling, permission was obtained from The Newhall Land and Farming Company, a wholly owned subsidiary of FivePoint Holdings, LLC (now Fivepoint)—owner of the Newhall Ranch population area, and Santa Monica Mountains Conservancy (owner of the Las Virgenes population area). Leaf and foliar samples were collected under California Department of Fish and Wildlife 2081 permits (CDFW 2014a; b).
Both seeds (common garden study) and foliage (genomic analysis and ploidy determination) were collected from all selected subpopulations at appropriate times based on plant phenology in 2014 and 2015. For seed collections, 15–20 individual plants were sampled from each of the 14 subpopulations. Collections were avoided from immediately adjacent plants and we maintained a three-meter distance among donor plants where possible to minimize likelihood of close genetic relationship. One involucral cluster was collected from individual plants and packaged separately by plant. These packages were kept in cool, dry conditions until transported to the nursery (California Botanic Garden facilities) for processing.
Seeds were cleaned using a combination of sieves, seed aspirators, and hand sorting to remove chaff and other inert material. Identity by maternal line (seed parent plant) was strictly maintained. Prior to initiating the common garden study, germination tests were conducted to determine an effective protocol. Based on those results, 40 seeds randomly selected from each subpopulation were placed on agar plates with a 0.5% agar solution and treated with a two-week cold, moist stratification period after which they were transferred to a germination chamber maintained at 11 h light cycle at 20 °C and 13 h dark cycle at 12 °C. All seedlings with cotyledons above the agar surface were sown directly into two-inch pots in a soil mixture of peat moss, cement sand, perlite, and osmocote (slow-release fertilizer).
Foliar samples were collected at the same 14 subpopulations earlier in the season to acquire fresh tissue. Foliage was collected from 20 plants per subpopulation; in some cases, several additional plants were sampled to allow a buffer or for protocol development. Similar to the sampling protocol for the seed collections, donor plants were at least three meters apart to the extent possible. A sample of the leaves on each donor plant were harvested and placed into a plant-specific and identified paper envelope and then into a cooler. Samples were kept cool and dry and delivered to the laboratory facility within a few days, at which point they were transferred to an ultralow freezer.
A previous unpublished pilot study testing the feasibility of using microsatellite loci for population genetic analyses of SFVS included C. staticoides, Turkish rugging—the only sympatric congener with SFVS. Significant genetic differentiation between Turkish rugging and SFVS was found (USDA Forest Service 2016). The two species are readily identified in the field as well; consequently, we did not include Turkish rugging samples for comparison.
Flow cytometry
Flow cytometry was used to check for evidence of ploidy variation in SFVS (USDA FS 2016). A consistent amount of leaf tissue (approximately 2.5 cm in length from leaves with oblanceolate to narrowly oblong shape) from ten individuals from each subpopulation was minced using double-edged safety razor blades in 1.5 mL Cystain UV Ploidy (DAPI) buffer (Sysmex-Partec, Görlitz, Germany) until the buffer solution took on a green hue. Samples were incubated in the dark at room temperature for 2.5 min before filtration though a 10 µm black CellTrics filter (Sysmex-Partec, Görlitz, Germany). A commercial variety of wheat was used as an internal standard to provide a relative measure and improve comparisons between spineflower assays. Wheat tissue was prepared and stained separately following the above protocol and 0.5 mL of wheat cell suspension was added to spineflower samples immediately prior to analysis. Samples were analyzed on a Partec PA Ploidy Analyzer (Sysmex-Partec, Görlitz, Germany), on the natural log scale (lower limit = 10, speed = 10, gain = 300). Because we were only interested in whether multiple ploidies were present, relative DNA content was estimated by quantifying the mean fluorescence of each sample and standardizing to the wheat mean fluorescence ([spineflower/wheat] × 100). Differences in ploidy were interpreted from the distribution of standardized relative DNA content values.
Common garden study
All potted seedlings (24–38 per subpopulation) were maintained in an outdoor shaded environment. Due to unusually high spring and summer temperatures, considerable seedling mortality occurred. However, seedlings from 13 of the 14 subpopulations survived (the Entrada subpopulation from Newhall Ranch had no survivors and was not included in the common garden study). In late May, 5–30 potted seedlings per subpopulation, based on survival, were randomly arranged on benches in the outdoor shaded environment. Measurements of specific growth and phenological traits were made from then until plants stopped growing. Some growth measurements were taken at two times: an early and late measurement (Table 2). When growth had ceased, all plants were cut at the soil surface and above-ground biomass fresh weight was measured. Dry weight was obtained by wrapping the plant material loosely in punctured foil, baking at 225 °C for four hours, and then weighing again. Seed production was not measured because this would have reasonably been influenced by the position of the plants on the bench (i.e., outcrossing with neighbors) and thus would have reflected environmental conditions as much or more than genetics.
Population genomics study
DNA extraction and RAD-seq library preparation
DNA was extracted from leaf tissue using the Qiagen DNEasy-96 (Qiagen, Venlo, Netherlands) Plant Kit following the liquid N protocol. We followed the manufacturer’s instructions but added a digestion with 80 mg of proteinase K for 30 min at 55 °C after the lysis incubation. DNA extracts were quantified with Fujifilm FLA 5100 fluorimager (Fujifilm Life Sciences, Tokyo, Japan).
Previous attempts to develop microsatellite markers for SFVS were unsuccessful (data not shown) so we performed restriction-site associated DNA sequencing (RAD-seq) to identify single nucleotide polymorphisms (SNPs) for population genetic analysis. RAD-seq libraries were prepared for 247 foliar samples following the protocol of Ali et al. (2016). Briefly, DNA was digested with the restriction enzyme Sbf1, which has an eight base pair recognition sequence. We ligated a unique barcode adapter to DNA from each sample which allowed us to combine multiple samples into a single Illumina HiSeq 4000 sequencing lane. Two libraries were submitted for a single lane of 100 bp paired end sequencing at the UC Davis DNA Technologies and Expression Analysis Core. Individuals from all sites were included in each sequencing lane to avoid lane effects.
Data analysis
Common garden study
Differences in phenotypic traits between populations and subpopulations were assessed using analysis of variance (ANOVA) with Tukey’s HSD tests for post-hoc pairwise comparisons. Prior to analyses, each trait was examined to verify the distributional assumptions of ANOVA. Two variables (DAYS2FLR and NUMFLE) did not meet these assumptions and were analyzed using Kruskal–Wallis non-parametric rank-based test with Dunn’s tests for post-hoc comparisons among subpopulations (p-values adjusted with the Benjamin-Hochberg method). Log-transformations were applied to normalize distributions and/or reduce heteroscedasticity as appropriate (DAYS2FL, HEIGHTE, LENGTHL, NUMFLE, WIDTHE, WIDTHL). To confirm that the position of plants during growth had no effects, variation in each trait was compared to flat and greenhouse position; we found no significant associations for either trait variable (all p-values > 0.05) and removed these from further consideration. Several datapoints represented “negative growth” and were removed from the final datasets for LENGTHL (n = 5), WIDTHL (n = 11), and HEIGHTL (n = 72), as these were assumed to represent measurement errors (e.g., some plants bent with growth thus reducing their dimensions). All analyses were performed in R v4.2.0 (R Core Team 2022) using the stats and FSA packages.
Population genomic analysis
De novo reference assembly and alignment
Sequence data were de-multiplexed and adapter sequences trimmed using custom scripts (0001_Demultiplex.sh). Reads from samples sequenced in more than one library were combined and the total number of reads enumerated (Table S1). A de novo assembly was generated with STACKS version 2.53 (Catchen et al. 2011, 2013) using 100 foliar samples that were neither extremely low nor extremely high in terms of read counts (0003_Stacks.sh). In brief, the STACKS program process_radtags was used to clean the sequence data, and ustacks was run on each sample to assemble loci for each individual. A catalog of available loci was generated with cstacks and samples matched back to the catalog with sstacks. To create a reference assembly, we used tsv2bam followed by gstacks. Reads were aligned to the resulting de novo assembly with the Burrows-Wheeler Aligner and the “mem” algorithm (bwa-mem version 0.7.17-r1188) (Li and Durban 2009) and filtered in SAMtools version 1.13 (Li et al. 2009) for proper pairing and PCR duplicates were removed (0004_Align.sh). Because some congenerics are known to be polyploid (e.g. C. orcuttiana; Hardham 1989), we identified paralogous loci that could bias population genetic analyses by subsampling alignments to approximately 60,000 reads each for even coverage and then using ANGSD version 0.930 (Korneliussen et al. 2014) and ngsParalog version 1.3.0 (0005_Subsample.sh, 0006_FindParalogs.sh). The output from realSFS was filtered based on an adjusted p-value generated by Bonferroni correction as suggested by the program author (0007_IdentifyingParalogs.Rmd). Any RAD locus containing paralogous sites was removed from subsequent analyses.
Population genomic analyses
We used ANGSD to call genotypes generating a PLINK-formatted file which was then converted to a variant call file (VCF). We used the following options with ANGSD: -doPlink 2 -doMajorMinor 1 -doMaf 1 -SNP_pval 1e-6 -minMapQ 10 -minQ 20 -minMaf 0.05 -doGeno 4 -doPost 1 -postCutoff 0.9. We also used the –minInd flag to require a SNP to be present in 90% of individuals and used the non-paralogous regions previously identified as a region file. The PLINK formatted file was converted to a VCF with PLINK version 1.90b6.14 (Purcell et al. 2007) and headings were changed to sample names with BCFtools version 1.10.1 (Danecek and McCarthy 2017).
Because SFVS exists in fragmented populations, is insect pollinated and self-fertilizes to varying degrees (Jones et al. 2009), inbreeding and selfing may arise at differing levels across populations. To test for the presence of inbreeding (f) in different populations of San Fernando Valley spineflower, identity disequilibrium was calculated as a proxy for f with the g2 statistic with the inbreedR package version 0.3.3 (Hoffman et al. 2014). The g2 statistic is robust to heterozygosity deficits due to factors other than inbreeding/selfing, such as genotyping errors or partial dominance (David et al. 2007). Nonetheless, the inbreedR authors advise that the most complete set of SNPs be used, unlinked SNPs may be preferable, genetically distinct populations be analyzed separately, and care be used in the inclusion of low minor allele frequency (MAF) SNPs (Stoffel et al. 2016). For the calculations of g2, we pruned the called SNP data set with BCFtools + prune (-l 0.9 -w 10,000) and to a proportion of missing data < 0.05. This pruned set of SNPS was filtered for each calculation of g2 to SNPs with a MAF > 0.1 and a proportion of missing data < 0.05. With the g2_snps function of inbreedR we specified 100 permutations and 100 bootstraps using other default settings of the function. From each mean and median value of g2 calculated, selfing (s), was calculated following (David et al. 2007). We calculated g2 and selfing (s) by pooling samples by population (ULVC and Newhall Ranch) and subpopulation.
We examined population genetic structure by first conducting a Principal Component (PC) analysis on the genotype calls within R. The VCF of genotypes was imported into R and converted to a genind object with the read.vcfR and vcfR2genind functions of the vcfR package version 1.13.0 (Knaus and Grünwald 2017). The genind object was converted to a matrix of allele data with the tab function of the adegenet package version 2.1.8, with missing data filled using mean values. We then used the dudi.pca function of the ade4 package version 1.7–20 for the PC analysis of genetic data. The first three axes of variation were retained to describe the largest patterns of genetic variation in SFVS.
To obtain an objective number of population genetic clusters (K), we conducted discriminant analysis of principal components (DAPC) on the genotype calls. Using the genind object previously generated, we used the find.clusters function of adegenet to find an optimal K based on the Bayesian information criterion (BIC). The genetic data were transformed into principal components and successive K-means clustering was conducted with increasing K. For each model, BIC was calculated to assess goodness of fit. A range of K values was examined using the default settings of find.clusters that is approximately the number of individuals divided by 10 (18 in this case).
To obtain pairwise FST and subpopulation nucleotide diversity (π) measurements, we excluded sites exhibiting signatures of inbreeding with PCAngsd version 0.982 (Meisner and Albrechtsen 2018, 2019). As PCAngsd uses genotype likelihoods (GLs), we created a Beagle-formatted file with ANGSD using the same specifications as the genotype calls except specifying that a GL file should be created (-doGlf 2) and major allele frequency was calculated (-doMaf 2). The resulting Beagle file was supplied to PCAngsd and a likelihood ratio test for inbred sites conducted (-inbreedSites and -minMaf 0.05 options used). Sites not exhibiting inbreeding were identified based on a threshold of 0.01 and saved for downstream analyses (commands in 0009-kinship-pca-admix.Rmd). For each subpopulation we generated site frequency spectra (SFS) with ANGSD and the realSFS subprogram (see script series 0011). The realSFS subprogram was then used to create 2-dimensional SFS and weighted FST measurements between each possible pair of subpopulations. Nucleotide diversity estimates for each contig were generated by using the realSFS saf2theta option followed by the thetaStat subprogram do_stat option.
Results
Flow cytometry
Flow cytometry was conducted successfully on 4–11 individuals per sampling site (N = 101). No evidence of ploidy variation was found among individuals or sites. All SFVS samples had a sample: wheat fluorescence ratio near 8.0.
Common garden study
At the population-level, all measures of reproductive-associated traits showed highly significant differences including DAYS2FLR (χ2(1) = 5.438, p = 0.0197), NUMFLE (χ2(1) = 14.162, p < 0.001), and NUMFLL (F(1) = 36.769, p < 0.001) (Fig. 3). The two populations also differed in regard to measurements of HEIGHTE (F(1) = 10.170, p = 0.0017), LENGTHL (F(1) = 3.929, p = 0.0493), NUMSTEML (F(1) = 4.945, p = 0.0276), and MASSDRY (F(1) = 6.935, p = 0.00935). The Las Virgenes population tended to flower sooner and more vigorously than Newhall and overall had greater biomass. There were no significant differences between populations in measurements of HEIGHTL, LENGTHE, NUMSTEME, or MASSWET (Table S2).
At the subpopulation-level, there were more traits that showed evidence of significant variation (p < 0.05) among the Las Virgenes subpopulations than among the Newhall Ranch subpopulations (7 traits vs. 4 traits, respectively) (Table S2). Within the Newhall Ranch population, traits that showed significant differences between subpopulations included NUMFLE, LENGTHE, WIDTHE, MASSDRY (p < 0.001 for all; Fig. 4). Within the Las Virgenes population, traits that showed significant differences between subpopulations included DAYS2FL, NUMFLE, NUMSTEME, LENGTHE, WIDTHE, MASSWET, and MASSDRY (p < 0.001 for all; Fig. 5).
Population genetic analysis
De novo reference assembly and alignment
The de novo reference contained 13,234 contigs and 182 spineflower samples aligned with greater than 79,000 reads after removal of PCR duplicates (Tables 1, S1). Forty-one samples were removed from the study due to an insufficient number of filtered reads. After screening for the presence of paralogs, 239 contigs were excluded for a total of 12,995 contigs with a mean length of 374 bp.
Population genetic analyses
A total of 10,073 SNPs were genotyped in the 182 spineflower samples. Evaluation of the g2 statistic (Table S3) indicated the presence of low to moderate selfing across subpopulations (Fig. 5). Newhall Ranch (3246 loci) and its subpopulations generally had higher estimates of selfing than ULVC (2864 loci) and its subpopulations (Figs. 5, 6). Selfing rates were variable among subpopulations at each location. LV 1 and 4 had higher selfing rates within ULVC while Entrada, Lion Canyon, and Valencia exhibited elevated selfing (s) in Newhall.
Principal component analysis of the variation present in the 10,073 SNPs shows a separation of Newhall Ranch, Las Virgenes (except LV6), and LV6 along PC 1 (3.66% of variance) (Fig. 7). Principal Component 2 aligns with separation within Las Virgenes (2.85% of variance) and PC 3 indicates separation within the Newhall Ranch subpopulations, with Entrada and Lion Canyon at the extremes (2.49% of variance). Successive K-means clustering while conducting DAPC using 150 PCs indicates a best fit of K = 3 based on BIC (Fig. 8A). These three clusters comprise (1) Newhall Ranch subpopulations, (2) Las Virgenes subpopulations excluding LV6, and (3) LV6 (Fig. 8B). Mean posterior probability assignments for each group are 1.00.
Population genetic diversity of San Fernando Valley spineflower. A Successive K-means clustering while conducting DAPC using 150 PCs, B Three clusters comprise (1) Newhall Ranch subpopulations, (2) ULVC subpopulations excluding LV6, and (3) LV6, C Pairwise FST excluding inbred SNPs, D Nucleotide diversity (π) is lowest in Lion Canyon, Entrada and LV6
Pairwise FST ranged from 0.05 to 0.25 with a mean of 0.14 (Figs. 8C, S2). Patterns of pairwise FST demonstrate the elevated distinctiveness of LV6 as well as the Entrada and Lion Canyon subpopulations. Median nucleotide diversity (π) is lowest in Lion Canyon, Entrada and Long Canyon (0.66, 0.727 and 0.785 respectively) (Fig. 8D). Highest median π is present in LV5, LV1 and Airport Mesa (0.961, 0.917 and 0.906 respectively).
Discussion
While the large number of rare and endangered plant species precludes the investment of focused genetic study afforded in model and agricultural species, well-designed studies with diverse methodologies can be efficient and effective tools in elucidating genetic information of conservation value. For example, combining approaches from population genomics and common garden studies can provide independent insights into the possibility of local adaptation scenarios (de Villemereuil et al. 2008). In this study, we used multiple approaches to evaluate the genetic status of the endangered San Fernando Valley spineflower. We confirmed with flow cytometry analysis that, although several spineflower congeners exhibit intraspecific chromosome count variation, the two remaining SFVS populations exhibit a single ploidy level. Intraspecific variation in chromosome number is an important consideration in rare plant management, particularly augmentation or reintroduction planning, because the progeny of interploid crosses often exhibit reduced F1 fertility (Ramsey and Schemske 1998). Chromosome counts are known for few listed species and even when known, those data aren’t often incorporated into recovery planning (Severns and Liston 2008).
We detected substantial levels of genetic diversity and low to moderate selfing rates that varied across subpopulations and that supports a mixed mating strategy. Significant genetic differentiation was estimated within and among populations using 10,073 SNP loci, and significant differences of phenotypic traits were observed between and within populations in our common garden experiment, which suggests that local adaptation may have occurred. This between-population structure is not unexpected given geographic distance and an intervening (although low) mountainous landscape with attendant impacts on pollinator distances.
The existence of significant genetic differentiation among populations and subpopulations (Figs. 7, 8) and median nucleotide diversity (π) values ranging from 0.660 to 0.961 (Fig. 8D) indicates that SFVS still retains substantial levels of genetic diversity, despite rarity and highly fluctuating population sizes. One contributing factor for observed levels of genetic diversity is the buffering effects of a persistent seed bank which can increase a subpopulation’s effective population size and allow multigenerational gene flow, reducing the rate of genetic drift (Honnay et al. 2008; Bradbury et al. 2016). Plue et al. (2017) found that a Campanula rotundifolia seed bank retained 80% of above-ground genetic diversity, as well as contained rare private alleles. In Clarkia springvillensis, a rare, narrowly endemic California annual plant, the seedbank contained higher genetic diversity than germinated plants (McCue and Holtsford 1998). Thus, the genetic diversity levels we report for SFVS likely represent an underestimate of standing variation in the taxon.
Interpreting the amount of genetic diversity in SFVS for conservation purposes would ideally be conducted in a context informed by genetic information from “comparable” species, including more common congeners if possible, and using similar genetic tools. In this case, we discovered little genetic information for congeneric species and few studies in rare, annual herbaceous plants using SNP loci.
A review of allozyme studies of 653 plant taxa that represented 449 species and 165 genera (Hamrick and Godt 1989) revealed that endemic species had less than half the genetic diversity of widespread species; annual species had lower genetic diversity than long-lived perennials.
Similarly, genetic diversity within populations showed some significant patterns—with selfing species and endemics being correlated with much lower levels of within-population genetic diversity than wind-pollinated and widespread species, respectively (Hamrick and Godt 1989). As an annual and with an apparently mixed mating system and highly restricted geographic range, SFVS may not be expected to have a high level of genetic diversity at the species or population levels—suggesting our study results are surprising. However, SNP data from other species, with both similar and differing life histories, would be required to determine whether levels of SVFS genetic diversity revealed here are consistent with the expectations of Hamrick and Godt (1989). Further, the expectation of lower genetic diversity in rare species may be a myth or at least an overly broad generalization. Gitzendanner and Soltis (2000) hypothesized that this relationship may be confounded by evolutionary history and, using a meta-analysis, demonstrated that neither genetic partitioning nor total levels of genetic diversity differed between rare and widespread congeners.
Genetic structure (among populations) has also been shown to be highly correlated with several traits, most notably breeding system and life form (Hamrick and Godt 1989). Selfing species showed much more population structure than did wind-pollinated, outcrossed species and annual species much more than long-lived woody perennials. These patterns may provide an expectation of considerable genetic structure for the SFVS, which indeed was the case—both between and within populations.
Evidence of population genetic differences and significant phenotypic differences between the Newhall and ULVC populations suggests both genetic drift and local adaptation are probably contributing to population structure within this taxon. Contemporary gene flow between Newhall and Las Virgenes was expected to be minimal given the distance between them (27 km), the intervening (low) mountain landscape, and land use conversion that has occurred in intervening habitat. Low landscape suitability can be a bigger driver of reduced gene flow and dispersal than geographic distance (Grasty et al. 2020; Hellwig et al. 2022). These factors would support genetic drift. Selective forces are also likely and local adaptation is common in plants (Oduor et al. 2016; Bucharova et al. 2019), even at fine geographic scales (Linhart and Grant 1996). In addition to the physical distance between the two populations—that can be a proxy for other environmental attributes—elevation for Las Virgenes subpopulations is generally higher than that at Newhall. Different selection pressures between locations were evidenced by the significantly larger size of Las Virgenes spineflower at several measures in the common garden experiment (Fig. 3). Current culture—monitoring and management activities at Newhall; relatively unmanaged landscape context at Las Virgenes—may also have an effect, currently or in the future. Determining the relative contributions of (neutral) genetic drift and natural selection to the observed population differences is challenging and not attempted.
Our study provided genetic evidence for a mixed mating strategy in San Fernando Valley spineflower. Selfing rates will influence how genetic diversity is partitioned within and among populations and subpopulations of this spineflower. Greater rates of selfing increase homozygosity and genetic drift and could accelerate a subpopulation’s genetic differentiation from other subpopulations (Glémin et al. 2006). Not unexpectedly, selfing rates were variable among subpopulations (Whitehead et al. 2018) and generally were lower within Las Virgenes than within most of the Newhall subpopulations. Within populations, Entrada and Lion Canyon, two subpopulations with relatively high genetic divergence from other Newhall Ranch subpopulations, have high selfing values (s), but Valencia, which also has a high s, is less genetically distinct. LV6 at Las Virgenes has a moderate selfing (s) value despite strong genetic divergence from all other subpopulations. Selfing rates can vary over time within and among subpopulations and it is possible that Valencia and LV6 exhibit higher selfing rates under different ecological or environmental conditions than those present in the year samples were collected (Whitehead et al. 2018). Selfing rates would need to be monitored across years to understand the role of selfing in genetic differentiation among spineflower populations. Additional research is needed to determine whether selfing or inbreeding negatively affects the fitness of individual plants.
Smaller geographic distances between subpopulations should increase the opportunity for gene flow but the existence of within-population genetic differentiation suggests genetic drift and/or local adaptation is occurring. Drift could result from isolation due to pollinator limitations and/or temporal variability in population size due to interannual variability in precipitation. In a study of pollinator visits to SFVS, it was observed that two ant species accounted for approximately 40% of all visits, with just three other species (European honey bees and two species of beetles) compromising another 35% of visits. Further, the four native pollinators tended to visit flowers on the same plant or nearby neighbors (Jones et al. 2009). Wingless pollinators and local pollination behavior would tend to limit gene flow.
Significant differences among subpopulations at phenotypic traits measured in our common garden experiment provide some evidence of local adaptation, but patterns of phenotypic variability didn’t always correspond to genetic differentiation. For example, the Lion Canyon subpopulation within Newhall was genetically divergent at SNP loci but was not significantly different at any measured phenotypic trait, while other subpopulations at Newhall and Las Virgenes with no strong genetic differentiation did differ significantly from others at one or more phenotypes. These results suggest that either our SNP set did not cover areas of the genome contributing to the phenotypic traits measured in the common garden study or that the basis of the trait variation is epigenetic or the result of maternal effects.
Other studies have shown that different methods of estimating genetic parameters—e.g., molecular or phenotypic/common garden—can reinforce patterns or provide contrasting interpretations. Some have shown that local adaptation can occur in the absence of genome-wide differentiation (Krohn et al. 2019; Hellwig et al. 2022). Marker diversity failed to provide evidence of genetic divergence that was demonstrated in a common garden study for a rare perennial herb, Arabis fecunda (McKay et al. 2001). Similarly, employing multiple approaches to assessing genetic diversity provided a more robust description of the amount and patterns of genetic diversity in another endangered California annual, Acanthomintha ilicifolia (DeWoody et al. 2018).
Subpopulation LV6 was an interesting outlier in our study. It exhibited high genetic divergence from other subpopulations, was identified as a separate genetic cluster by DAPC, and exhibited high pairwise FST values. It also differed significantly in reproductive traits, flowering 5–10 days earlier than other subpopulations at Las Virgenes and possessing more flowers early in the inflorescence season. In combination, this evidence suggests that both drift and adaptive processes have contributed to the uniqueness of LV6. Earlier flowering time could limit gene flow between LV6 and other subpopulations and promote inbreeding and/or selfing, although the g2 and selfing (s) values for LV6 weren’t remarkable when compared to other subpopulations. LV6 is at the eastern edge of SFVS’s current distribution in Las Virgenes, and is the farthest from its nearest neighboring subpopulation (LV5—0.6 km) than any of the others. Thus gene flow—if mainly mediated by short-distance pollinators—may be more limiting than among the other subpopulations. If environmental conditions on the eastern edge are less optimal, LV6 may germinate more intermittently, temporally isolating it from other Las Virgenes subpopulations. Temporal genetic structure was shown to exceed spatial genetic structure in an allozyme study of the slender-horned spineflower (Dodecahema leptoceras; Ferguson and Ellstrand 1999). Also, LV6 is found on a wash that is at lower elevation from other subpopulations, potentially limiting pollinator access. Different soil type (San Andreas silty loam) at LV6 relative to other sampled subpopulations could be a selective factor. Finally, it is possible that LV6 represents the remnant of another spineflower population that is geographically adjacent to but genetically distinct from the other Las Virgenes subpopulations. Additional study of SFVS in Las Virgenes Open Space Preserve is required to determine the mechanisms driving divergence between LV6 and other subpopulations.
Conservation implications
With only two extant natural populations, conservation measures for this endangered taxon include attempting to establish new populations and managing the existing populations (USFWS and The Newhall Land and Farming Company 2017). The finding of significant genetic differences in both quantitative traits and at molecular levels, as well as known environmental differences, between Newhall and Las Virgenes suggests that they should be managed as separate populations. While some natural gene flow could still occur between the populations, the short travel distances associated with the most likely seed and pollinator dispersers suggest that any gene flow could be easily swamped by local adaptation.
When attempting to establish new populations, the observed genetic structure suggests using seed from the subpopulation with the closest match in environmental conditions. Although the selective pressures driving potential adaptive differences among subpopulations are unknown, there is sufficient evidence of local adaptation to select recipient sites not only for suitability at the taxon level, but for the closest possible subpopulation site match. If the recipient site for reintroduction or establishment has environmental conditions that reflect more than one extant subpopulation, and as long as the recipient site is sufficiently distant from extant subpopulations to avoid most gene flow, regional admixture provenancing, or drawing seeds from two or more subpopulations, might be considered (Bucharova et al. 2019). The mixing of seeds/plants from multiple subpopulations would capture higher levels of genetic diversity than drawing from a single source and may increase the likelihood of the establishment of new subpopulations (Bucharova et al. 2019; Höfner et al. 2022). Introducing individuals from multiple subpopulations broadens the suite of phenotypes on which natural selection at a site can act. Although using admixtures is risky, and success (persistence) would not be evident for a long time, greater risks can be assumed in establishing new populations (that are well removed from extant populations) than in measures that could genetically impact extant populations.
Regarding genetic management of extant populations, we found no evidence that they were genetically depauperate and, as such, genetic interventions (such as direct seeding) would not appear to be warranted. We also note that the soil seed bank reasonably represents a genetic diversity reservoir. We recommend that managers support extant levels of genetic diversity by maintaining existing mechanisms of gene flow among and within subpopulations and natural reproduction. The most obvious means to do so is by supporting appropriate pollinator (and seed dispersing) populations including monitoring for and managing as needed, invasive Argentine ants (Linepithema humile). This guidance is further supported by a meta-analysis of relationships among genetic diversity, fitness, and population size with plant features including rarity and life history traits which suggested that reduced pollinator activity generally contributes to reduced fitness of plants in small populations of rare species (Leimu et al. 2006).
Genetic diversity and structure should be reassessed periodically (e.g., decadal) to allow managers to identify changes that may have management implications. Although recommendations for genetic monitoring seem to focus on translocated and/or mixed gene pool populations to reveal effects, for example, from outbreeding depression and/or natural selection (Van Rossum et al 2020; Albrecht and Edwards 2020; Van Rossum and Hardy 2022), this is also a prudent practice for small populations of a vulnerable taxon. Further, management practices could affect genetic diversity over time. In addition, understanding how long seeds are viable in the seed bank and how seeds move across the landscape will help managers understand the likelihood of extirpation and natural recolonization after repeated dry years. Ferguson and Ellstrand (1999) reported that germination in the slender-horned spineflower can occur several years after the most recent seed production but viability of the seed bank over time has yet to be characterized in the San Fernando Valley spineflower.
Small populations, such as individual San Fernando Valley spineflower subpopulations, are susceptible to genetic diversity loss and may not be able to adapt to rapid climate change, although the annual life form is advantageous for rapid adaptation. For example, this taxon exhibits variable germination rates with higher germination in wet years than in drier years (Jones et al. 2009). The increased frequency of drought in California may increase temporal variation in subpopulation size and fitness and this stochasticity may accelerate genetic drift. Maintaining adequate levels of pollination and supporting pollinators (both known pollinators and a diversity of species) is essential towards maintaining genetic diversity. Overall, this taxon—based on one-time samples but multiple lines of evidence—appears to harbor substantial levels of genetic diversity with significant structuring that is suggestive of local adaptation, accompanied by drift. Given its undetected persistence across decades and soil seed bank, this San Fernando Valley spineflower appears to have an inherent robustness that may be best supported by attempting to establish new populations while avoiding anthropogenic influences on extant populations more so than any direct intervention at this time.
Data availability
Raw sequence reads generated in this study can be found in the NCBI Sequence Read Archive under accession PRJNA873787. Scripts used in analysis of RADseq data are available at https://github.com/MacCampbell/spineflower.
References
Albrecht MA, Edwards CE (2020) Genetic monitoring to assess the success of restoring rare plant populations with mixed gene pools. Mol Ecol 29:4037–4039. https://doi.org/10.1111/mec.15658
Ali OA, O’Rourke SM, Amish SJ, Meek MH, Luikart G, Jeffres C, Miller MR (2016) RAD capture (Rapture): flexible and efficient sequence-based genotyping. Genetics 202:389–400. https://doi.org/10.1534/genetics.115.183665
Bay RA, Harrigan RJ, Underwood VL, Gibbs HL, Smith TB, Ruegg K (2018) Genomic signals of selection predict climate-driven population declines in a migratory bird. Science 359:83–86. https://doi.org/10.1126/Science.Aan4380
Bradbury D, Tapper SL, Coates D, McArthur S, Hankinson M, Byrne M (2016) The role of fire and a long-lived soil seed bank in maintaining persistence, genetic diversity and connectivity in a fire-prone landscape. J Biogeogr 43:70–84. https://doi.org/10.1111/jbi.12601
Bucharova A, Bossdorf O, Hölzel N, Kollmann J (2019) Mix and match: regional admixture provenancing strikes a balance among different seed-sourcing strategies for ecological restoration. Cons Gen 20:7–17. https://doi.org/10.1007/s10592-018-1067-6
California Department of Fish and Game (CDFG) (2002) Report to the fish and game commission on the status of San Fernando Valley spineflower (Chorizanthe parryi var. fernandina). pp 3 + appendices
California Department of Fish and Wildlife (CDFW) (2014a) California endangered species act scientific, educations, or management permit No. 2081(a)-14–007-RP. San Fernando Valley Spineflower genetics study (Upper Las Virgenes Canyon Open Space Preserve), PI Deborah Rogers, Center for Natural Lands Management, May 16, 2014
California Department of Fish and Wildlife (CDFW) (2014b) California endangered species act scientific, educations, or management permit No. 2081(a)-14-010-RP. San Fernando Valley Spineflower genetics study (Newhall Ranch), PI Deborah Rogers, Center for Natural Lands Management, July 25, 2014
California Natural Diversity Database (CNDDB) (2022a). RareFind 5 [Internet]. California Department of Fish and Wildlife Version 5
California Natural Diversity Database (CNDDB) (2022b) State and federally listed endangered, threatened, and rare plants of California, October 2022. California Department of Fish and Wildlife Sacramento, CA
California Native Plant Society, Rare Plant Program (CNPS) (2022) Rare plant inventory (online edition, v9–01 1.5). https://www.rareplants.cnps.org. Accessed 27 Oct 2022
Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH (2011) Stacks: building and genotyping loci de novo from short-read sequences. Genes Genomes Genet 1:171–182. https://doi.org/10.1534/g3.111.000240
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140
Danecek P, McCarthy SA (2017) BCFtools/csq: haplotype-aware variant consequences. Bioinformatics 33:2037–2039. https://doi.org/10.1093/bioinformatics/btx100
David P, Pujol B, Viard F, Castella V, Goudet J (2007) Reliable selfing rate estimates from imperfect population genetic data. Mol Ecol 16:2474–2487. https://doi.org/10.1111/j.1365-294X.2007.03330.x
de Villemereuil P, Gaggiotti OE, Mouterde M, Till-Bottraud I (2008) Common garden experiments in the genomic era: new perspectives and opportunities. Heredity 116:249–254. https://doi.org/10.1038/hdy.2015.93
Dewoody J, Rogers DL, Hipkins VD, Endress BA (2018) Spatially explicit and multi-sourced genetic information is critical for conservation of an endangered plant species, San Diego thornmint (Acanthomintha ilicifolia). Cons Gen 19:893–907. https://doi.org/10.1007/s10592-018-1062-y
Diffenbaugh NS, Swain DL, Touma D (2015) Anthropogenic warming has increased drought risk in California. Proc Natl Acad Sci USA 112:3931–3936. https://doi.org/10.1073/pnas.1422385112
Dudek (2010) Spineflower conservation plan. The Newhall Land and Farming Company, California
Ferguson NJ, Ellstrand NC (1999) Assessment of seed bank buffering of genetic change in Dodecahema leptoceras (slender-horned spineflower), Contract No. FG-7642R5. California Department of Fish and Game, Long Beach, p 36
Gitzendanner MA, Soltis PS (2000) Patterns of genetic variation in rare and widespread plant congeners. Am J Bot 87:783–792. https://doi.org/10.2307/2656886
Glémin S, Bazin E, Charlesworth D (2006) Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc Royal Soc Biol 273:3011–3019. https://doi.org/10.1098/rspb.2006.3657
Grasty MR, Thompson PG, Hendrickson EC, Pheil AE, Cruzan MB (2020) Fine-scale habitat heterogeneity and vole runways influence seed dispersal in Plagiobothrys nothofulvus. A J Bot 107:413–422. https://doi.org/10.1002/ajb2.1433
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding, and genetic resources. Sinauer Associates, Inc., Sunderland, pp 44–64
Hardham CB (1989) Chromosome numbers of some annual species of Chorizanthe and related genera (Polygonaceae: Eriogonoideae). Phytologia 66:89–94
Hellwig T, Abbo S, Ophir R (2022) Drivers of genetic differentiation and recent evolutionary history of an Eurasian wild pea. J Biogeog 49:794–808. https://doi.org/10.1111/jbi.14274
Hickman JC (1993) The Jepson manual: higher plants of California. University of California Press, Berkeley
Hoffman JI, Simpson F, David P, Rijks JM, Kuiken T, Thorne MAS, Lacy RC, Dasmahapatra KK (2014) High-throughput sequencing reveals inbreeding depression in a natural population. Proc Natl Acad Sci USA 111:3775–3780. https://doi.org/10.1073/pnas.1318945111
Höfner J, Klein-Raufhake T, Lampei C, Mudrak O, Bucharova A, Durka W (2022) Populations restored using regional seed are genetically diverse and similar to natural populations in the region. J Appl Ecol 59:2234–2244. https://doi.org/10.1111/1365-2664.14067
Honnay O, Bossuyt B, Jacquemyn H, Shimono A, Uchiyama K (2008) Can a seed bank maintain the genetic variation in the above ground plant population? Oikos 117:1–5. https://doi.org/10.1111/j.2007.0030-1299.16188.x
IPCC (2022) Summary for policymakers. In: Pörtner HO, Roberts DC, Tignor M, Poloczanska ES, Mintenbeck K, Alegría A, Craig M, Langsdorf S, Löschke S, Möller V, Okem A, Rama B (eds) Climate change 2022: impacts, adaptation and vulnerability: contribution of working group II to the sixth assessment report of the intergovernmental panel on climate change. Cambridge University Press, Cambridge, pp 3–33. https://doi.org/10.1017/9781009325844.001
Jones CE, Shropshire FM, Taylor-Taft LL, Walker SE, Song LC Jr, Atallah YC, Allen RL, Sandquist DR, Luttrell J, Burk JH (2009) Reproductive biology of the San Fernando Valley spineflower, Chorizanthe parryi var. fernandina (Polygonaceae). Madroño 56:23–42. https://doi.org/10.3120/0024-9637-56.1.23
Jones CE, Atallah YC, Shropshire FM, Luttrell J, Walker SE, Sandquist DR, Allen RL, Burk JH, Song LC Jr (2010) Do native ants play a significant role in the reproductive success of the rare San Fernando Valley spineflower, Chorizanthe parryi var. fernandina (Polygonaceae)? Madroño 57:161–169. https://doi.org/10.3120/0024-9637-57.3.161
Kelley M (2019) Adaptation to climate change through genetic accommodation and assimilation of plastic phenotypes. Phil Trans R Soc B 374:20180176. https://doi.org/10.1098/rstb.2018.0176
Knaus BJ, Grünwald NJ (2017) vcfr: a package to manipulate and visualize variant call format data in R. Mol Ecol Resour 17:44–53. https://doi.org/10.1111/1755-0998.12549
Korneliussen T, Albrechtsen A, Nielsen R (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinform 15:356. https://doi.org/10.1186/s12859-014-0356-4
Krohn AR, Diepeveen ET, Bi K, Rosenblum EB (2019) Local adaptation does not lead to genome-wide differentiation in lava flow lizards. Ecol Evol 9:6810–6820. https://doi.org/10.1002/ece3.5231
Leimu R, Mutikainen P, Koricheva J, Fischer M (2006) How general are positive relationships between plant population size, fitness and genetic variation? J Ecol 94:942–952. https://doi.org/10.1111/j.1365-2745.2006.01150.x
Li H, Durban R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinform 2:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinform 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Linhart YB, Grant MC (1996) Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Evol Syst 27:237–277. https://doi.org/10.1146/annurev.ecolsys.27.1.237
McCue K, Holtsford T (1998) Seed bank influences on genetic diversity in the rare annual Clarkia springvillensis (Onagraceae). Am J Bot 85:30–36
McGraw JM (2018) Newhall Ranch San Fernando Valley spineflower habitat characterization. Jodi McGraw Consulting Draft Report, January 2018
McKay JH, Bishop JG, Lin JZ, Richards JH, Sala A, Mitchell-Olds T (2001) Local adaptation across a climatic gradient despite small effective population size in the rare sapphire rockcress. Proc R Soc Lond B 268:1715–1721
Meisner J, Albrechtsen A (2018) Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210:719–731. https://doi.org/10.1534/genetics.118.301336
Meisner J, Albrechtsen A (2019) Testing for Hardy-Weinberg equilibrium in structured populations using genotype or low-depth next generation sequencing data. Mol Ecol Resour 19:1144–1152. https://doi.org/10.1111/1755-0998.13019
Oduor AMO, Leimu R, van Kleunen M (2016) Invasive plant species are locally adapted just as frequently and at least as strongly as native plant species. J Ecol 104:957–968
Plue J, Vandepitte K, Honnay O, Cousins SAO (2017) Does the seed bank contribute to the build-up of a genetic extinction debt in the grassland perennial Campanula rotundifolia? Ann Bot 120(3):373–385. https://doi.org/10.1093/aob/mcx057
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
R Core Team (2022) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Evol Syst 29:467–501. https://doi.org/10.1146/annurev.ecolsys.29.1.467
Razgour O, Forester B, Taggart JB, Bekaert M, Juste J, Ibáñez C, Puechmaille SJ, Novella-Fernandez R, Alberdi A, Manel S (2019) Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1820663116
Reveal JL, Hardham CB (1989) A revision of the annual species of Chorizanthe (Polygonaceae: Eriogonoideae). Phytologia 66:98–198
Sapphos (Sapphos Environmental Inc) (2001) An investigation of the San Fernando Valley spineflower for the Ahmanson Land Company. Sapphos Environmental Inc, Pasadena
Savolainen O, Lascoux M, Merilä J (2013) Ecological genomics of local adaptation. Nat Rev Genet 14:807–820. https://doi.org/10.1038/nrg3522
Severns PM, Liston A (2008) Intraspecific chromosome number variation: a neglected threat to the conservation of rare plants. Cons Bio 22:1641–1647. https://doi.org/10.1111/j.1523-1739.2008.01058.x
Stoffel MA, Esser M, Kardos M, Humble E, Nichols H, David P, Hoffman JI (2016) inbreedR: an R package for the analysis of inbreeding based on genetic markers. Method Ecol Evol 7:1331–1339. https://doi.org/10.1111/2041-210X.12588
United States Department of Agriculture Forest Service (USDA FS) and National Forest Genetics Laboratory (NFGEL) (2016) Genetic studies of Chorizanthe parryi var. fernandina (S. Watson) Jeps. San Fernando Valley spineflower. Report of Project Development, NFGEL Project 299, Hipkins V, DeWoody J
United States Fish and Wildlife Service (USFWS) (1999) Endangered and threatened wildlife and plants; review of plant and animal taxa that are candidates or proposed for listing as endangered or threatened; annual notice of findings on recycled petitions; annual description of progress on listing actions; proposed rule. Fed Reg 64:57534–57547
United States Fish and Wildlife Service (USFWS) (2004) Species assessment and listing priority assignment form. Chorizanthe parryi var. fernandina. p 12
United States Fish and Wildlife Service (USFWS) (2016) Threatened species status for Chorizanthe parryi var. fernandina (San Fernando Valley Spineflower); Proposed rule. Fed Reg 81:63454–63466
United States Fish and Wildlife Service (USFWS) and The Newhall Land and Farming Company (2017) Candidate conservation agreement for the San Fernando Valley spineflower. https://ecos.fws.gov/docs/plan_documents/tcca/tcca_2254.pdf
Van Rossum F, Hardy OJ (2022) Guidelines for genetic monitoring of translocated plant populations. Cons Bio 36:e13670. https://doi.org/10.1111/cobi.13670
Van Rossum F, Hardy OJ, Le Pajolec S, Raspé O (2020) Genetic monitoring of translocated plant populations in practice. Mol Ecol 29:4040–4058. https://doi.org/10.1111/mec.15550
Whitehead MR, Lanfear R, Mitchell RJ, Karron JD (2018) Plant mating systems often vary widely among populations. Front Ecol Evol. https://doi.org/10.3389/fevo.2018.00038
Wright S (1943) Isolation by distance. Genetics 28:114–138. https://doi.org/10.1093/genetics/28.2.114
Acknowledgements
The authors acknowledge and appreciate funding provided by The Newhall Land and Farming Company, a wholly owned subsidiary of FivePoint Holdings, LLC; Agreement # 82085262 OL and by the Center for Natural Lands Management. We thank Drs. Anuja Parikh and Nathan Gale for invaluable and delightful assistance in locating subpopulations and conducting seed and foliage collections. We thank Drs. Jennifer DeWoody and Valerie Hipkins (NFGEL, USDA Forest Service, USDA) for flow cytometry ploidy determination and initial greenhouse culture studies. We thank the property owners and conservation managers (FivePoint Communities-Valencia [aka Newhall Ranch]—owned and managed by Newhall/FivePoint; Upper Las Virgenes Canyon Open Space Preserve—owned by Santa Monica Mountains Conservancy and managed by Mountains Recreation and Conservation Authority) for allowing us access for sample collections and for protecting the wild populations; the California Department of Fish and Wildlife for protecting the wild populations and permitting their sampling for this study. Thanks also to Alisha Goodbla for preparing RADseq libraries for sequencing. This paper is more robust in its content and eloquent in its presentation as a result of comments from the anonymous reviewers.
Funding
Funding was provided by The Newhall Land and Farming Company, a wholly owned subsidiary of FivePoint Holdings, LLC; Agreement # 82085262 OL and Center for Natural Lands Management.
Author information
Authors and Affiliations
Contributions
This research study was conceived by DLR. with contributions from ADS. All authors contributed to methodology and interpretations. CB and DLR performed lab and field work. Data analyses were performed by MAC and MAL with contributions from DLR and LKW. Authors ADS, DLR, MAC and MAL contributed to the original draft preparation and ADS, DLR, LKW, MAC and MAL contributed to review. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
Authors ADS, CB, LKW, and MAC have no conflicts of interest to declare that are relevant to the content of this article. Financial interests: Authors DLR and MAL are employed by and receive salary from the Center for Natural Lands Management.Non-financial interests: Authors DLR and MAL are employees of the Center for Natural Lands Management, which holds a Conservation Easement over lands owned by The Newhall Land and Farming Company, a wholly owned subsidiary of FivePoint Holdings, LLC; it is intended that ownership of this property be transferred to a Joint Powers Authority in the near future.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rogers, D.L., Washburn, L.K., Birker, C. et al. Genomic and common garden data reveal significant genetic differentiation in the endangered San Fernando Valley spineflower Chorizanthe parryi var. fernandina. Conserv Genet (2024). https://doi.org/10.1007/s10592-024-01610-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10592-024-01610-0