Abstract
Elasmobranchs (sharks and rays) are the most threatened marine vertebrates, particularly in tropical and subtropical areas. Their population status is often poorly understood due to insufficient information. Despite reportedly harbouring critical elasmobranch habitats, the Banc d’Arguin National Park (PNBA) in Mauritania lacks comprehensive and updated information on the diversity of elasmobranch species in the area. We developed a baseline inventory based on morphological and molecular identification and metabarcoding. DNA barcoding of tissue samples from elasmobranch processing sites and freshly sampled specimens was used to build a genetic reference database of local elasmobranch species. The richness and diversity of species in the PNBA were described via metabarcoding of seawater eDNA samples using an elasmobranch-specific assay and our reference database. We detected 27 species, including 12 new species records for the PNBA. We further uncover potentially undescribed species of Gymnura and Torpedo, while taxonomic corrections are noted for previously reported species. In particular, the reportedly abundant Mustelus mustelus was absent from tissue and eDNA samples, while M. punctulatus was detected instead. Taxa that have anecdotally become regionally extinct or rare (e.g., sawfishes, wedgefishes, lemon sharks) were not detected, highlighting local species diversity shifts within the last few decades. Results show that 67.9% of elasmobranch species in the PNBA are threatened with extinction according to the IUCN Red List of Threatened Species. This study emphasises the importance of taxonomic identification in support of species management and provides a baseline to inform future studies and conservation measures to avoid further species losses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Elasmobranchs (sharks and rays) are second only to Amphibians as the most threatened group of vertebrates, with an estimated 37% of species at high risk of extinction (Dulvy et al. 2021a). Overexploitation due to both intentional and unintentional catches (i.e. bycatch) from fishing is the leading cause of global elasmobranch population declines (Dulvy et al. 2021a), as biological traits such as slow growth, late maturity and low fecundity put them at higher risk of unsustainable exploitation relative to other commercially fished species (Simpfendorfer and Kyne 2009; Erhardt and Weder 2020). However, elasmobranchs continue to be extracted in large numbers (FAO 2014; Dulvy et al. 2021a), a practice that is primarily fuelled by the trade in fins and meat (Dent and Clarke 2015; Fields et al. 2018; Pincinato et al. 2022). Implementing fishing and trade regulations is essential for elasmobranch conservation and should be supported by understanding species diversity and trends in abundance and distribution.
Baseline data on species diversity and abundance from many global fisheries are often poor due to unreported landings or unresolved taxonomy of reported catches (Burgess et al. 2005; FAO 2014; Cashion et al. 2019). Accurate species-specific data are important for effective conservation and management of elasmobranchs (FAO 2014). Erroneous species identifications can lead to biased data on size, maturity, and abundance (Smart et al. 2016), directly affecting fisheries management (Burgess et al. 2005). However, some elasmobranch species can be difficult to distinguish morphologically, even by trained observers (Tillett et al. 2012; Smart et al. 2016).
In addition to accurate taxonomy, effective conservation requires species distribution information. Traditional survey methods such as Underwater Visual Census (UVC), Baited Remote Underwater Visual Surveys (BRUVS), and fisheries-independent surveys can be time, effort, and resource-intensive and are often inefficient at detecting rare and elusive species (Thomsen et al. 2012; Simpfendorfer et al. 2016; Boussarie et al. 2018; Budd et al. 2021). In recent years, environmental DNA (eDNA) metabarcoding has emerged as a non-invasive and cost-efficient technique to infer species composition. However, eDNA metabarcoding is strongly influenced by the marker selected and by the completeness of reference databases (Marques et al. 2021). While the COI gene has been the standard barcoding marker for animals (Hebert et al. 2003a, b), the 12S gene has shown higher specificity with more taxa identified for certain taxonomic groups (Collins et al. 2019; Zhang et al. 2020). The 12S elasmobranch-specific primers (Miya et al. 2015) have been generally effective at accurately reflecting species composition, in some cases more efficiently than traditional methods (Yamamoto et al. 2017; Lafferty et al. 2021; Mariani et al. 2021).
Many threatened elasmobranch species occur in Mauritania, where landings of sharks and rays are high (FAO 2023). Mauritania constitutes a natural boundary for southern and northern distribution limits of various species, leading to unique assemblages of taxa from different biogeographic affinities and supporting the presence of up to 115 species of sharks, rays, and chimaeras (Last et al. 2016; Ebert et al. 2021). This is comparable with regions of high species richness such as Peru or Madagascar (Cornejo et al. 2015; Fricke et al. 2018). Bordering the northern coast of Mauritania, the National Park of the Banc d’Arguin (PNBA), West Africa’s largest marine protected area (MPA), likely plays a vital role as feeding and reproductive grounds for elasmobranchs regionally (Valadou et al. 2006; Trégarot et al. 2018).
Following a surge of elasmobranch targeted fisheries in West Africa in the 1980s, regional populations of sharks and rays were reported as overexploited by the end of the century (Diop and Dossa 2011), including in the Banc d’Arguin. In 2000, a law was passed (Loi n° 2000.024) regulating small-scale fisheries within the PNBA by giving exclusive fishing rights to local fishers (Imraguen) based solely on subsistence fishing, defined as keeping at least 50% of catches for local consumption (Loi N°2000-025 du Code des Pêches de Mauritanie). Additionally, the PNBA administration and the local fishing community jointly agreed on regulatory measures in 2003 that resulted in a moratorium on most targeted elasmobranch fishing within the PNBA (Ducrocq et al. 2004; FAO 2018). However, fishing pressure on elasmobranchs has continued to increase (Failler et al. 2009; Barham et al. 2011; Trégarot et al. 2020) despite not being consumed locally (Jabado, unpublished data), but rather due to the lucrative nature of their products and insufficient local monitoring and enforcement capacities (FAO 2018). Anecdotal evidence suggests regional extinction for some species such as sawfishes (Pristidae) (Leeney and Downing 2016) and the endemic false shark ray, Rhynchorhina mauritaniensis (Kyne et al. 2020), while others have become increasingly rare, such as wedgefishes (Rhinidae) or lemon sharks (Negaprion brevirostris) (Diop and Dossa 2011). Many other species that are commonly landed in the PNBA (e.g., the scalloped hammerhead, Sphyrna lewini or the blackchin guitarfish, Glaucostegus cemiculus) face a high risk of extinction globally (Kyne and Jabado 2019; Rigby et al. 2019; Trégarot et al. 2020). Therefore, obtaining updated, accurate information on local elasmobranch diversity and distribution is increasingly important to inform decision-makers and policymakers across the region.
This study describes the current elasmobranch species diversity in the largest National Park of Atlantic Africa, the PNBA. To achieve this, we (a) created a DNA barcode reference database of local elasmobranch species to cross-reference with eDNA samples and to validate and correct taxonomy based on morphological and genetic assessments; (b) sampled seawater eDNA at different sites within the PNBA and applied metabarcoding to estimate the elasmobranch species present using the database produced in a); and (c) contrasted species diversity previously reported or suspected in the PNBA with our present estimate of species diversity confirmed visually, through DNA barcoding, or detected in eDNA samples.
Materials and methods
Sampling location
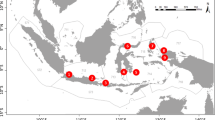
Mauritania is located in western, sub-Saharan Africa, bordering the central-eastern Atlantic Ocean. The Parc National du Banc d’Arguin (PNBA; Fig. 1) occupies over 180 km of Mauritanian coastline, encompassing 12.000 km² split between land and sea. The Banc d’Arguin is a large shallow water bay composed mainly of intertidal sandbanks, mudflats, intricate channels, and several dispersed islands (Wolff et al. 1993). It is fuelled by cold, nutrient-rich water from a permanent upwelling zone in the north, which is the primary driver of high regional productivity (Valdés and González 2015). The PNBA is inhabited by the Imraguen indigenous artisanal fisher community (Boulay 2013).
Sampling methods
To build a comprehensive reference DNA barcode database of local elasmobranch species, a total of 217 tissue samples were obtained, mostly from the national elasmobranch processing and trading sites Bountiya (n = 159) and Blaouakh (n = 35) between October 2020 and April 2021 (Fig. 1). These sites were chosen because national elasmobranch landings from artisanal or industrial fisheries are primarily transported and processed there, allowing access to the largest selection of local species for genetic sampling. Additional samples were collected opportunistically from specimens landed at Iwik (n = 17), an individual found dead in Arkeiss, and live animals (n = 5) caught during fisheries-independent surveys in the PNBA. Tissue samples (1 cm2) were taken from pelvic fins, to avoid sampling the same specimen twice. In cases where pelvic fins were absent, any available fin tissue from the right side of the body was collected. Depending on the species abundance at sampling (identified in situ based on morphology), three to five individuals were sampled per species. Samples were stored in 96% ethanol and kept at 4 °C upon arrival at the laboratory until DNA extraction.
Photographic vouchers were retained for each sampled individual to revise initial species identification using morphological identification keys (Last et al. 2016; Ebert et al. 2021), except for samples 159–166 and 401–404 (Online Resource 1). The former were freshly landed rays at Iwik being prepared for transportation, and the latter were live specimens released immediately after sampling, placing a time constraint on the sampling process in both cases.
DNA barcoding
DNA barcoding was used in the molecular identification of the specimens, and the resulting data was used to create a local reference sequence database needed for the eDNA metabarcoding of seawater samples. During both stages (i.e., DNA barcoding and eDNA metabarcoding), hygiene control protocols were strictly enforced to prevent contamination (i.e., lab working spaces and equipment were sterilised and single-use filtered pipette tips were used).
Genomic DNA (gDNA) was extracted from tissue samples using a NaCl protocol with a single ethanol washing step (Sambrook and Russell 2001). DNA quality was assessed using 0.8% agarose gel electrophoresis stained with Gelred (Biotium, Inc), and quantified in Nanodrop 1000 (ThermoFisher). The MiFish-E universal primer pair (Miya et al. 2015) was used for PCR amplification of a small region (~ 200 bp) of the 12S mitochondrial gene. Each PCR reaction included 2X Colorless GoTaq Flexi Buffer, 8 mM MgCl2, 320µM dNTP, 0.2 µM of each primer, 1.25 U GoTaq G2 Flexi DNA polymerase (Promega), and 2 µl of gDNA (1–5 ng/µl) on a 25 µl volume. PCR conditions consisted of an initial 2-minute denaturation phase at 95 °C followed by 30 cycles of 30 s of denaturation at 95 °C, 30 s of annealing at 55 °C and 1 min of extension at 72 °C with a final extension phase of 5 min at 72 °C. They were subsequently run on a GeneAmp® PCR System 9700 Thermal Cycler (Applied Biosystems). A subset of samples (n = 76) with lower amplification success was re-extracted with a slightly amended protocol to yield cleaner DNA by soaking them overnight in autoclaved Milli-Q water to remove excess salts and adding two additional ethanol and a final isopropanol washing step. Final PCR products were bi-directionally sequenced at CCMAR’s Sequencing Platform with an Applied Biosystems 3130xl Genetic Analyzer, BigDye®Terminator V3.1 chemistry, and POP7 polymer.
Sequence ends were trimmed, and the quality of pairwise assembled sequences was assessed and checked for consistency using Geneious Prime® 2021.1.1. Samples were identified using the Basic Local Alignment Research Tool (BLAST), comparing the newly generated sequences to those deposited in GenBank (National Centre for Biotechnology Information - NCBI) based on similarity percentages. Only matches above 98% were considered for species-level identification, and sequences with matches below 98% or where no reference sequence exists for a non-identified species were noted and preliminarily identified based on morphological assessments. In cases where a given sequence had a similarity above 98% or 100% for multiple taxa, the hit with the highest % similarity or the highest likelihood based on morphology and distribution was kept, respectively.
To improve taxonomic resolution for unresolved species, partial regions of mitochondrial COI (~ 650 bp) and NADH2 (~ 1050 bp) genes were targeted for a subset of samples (COI = 95, NADH2 = 23) using the universal primer pair Fish-F1 & R1 or Fish-F2 & R2 for COI (Ward et al. 2005) and the universal primer pair ILEM & ASNM (Naylor et al. 2012) and genus-specific primers for Mustelus spp. (Naylor et al. 2005) for NADH2 (Online Resource 2).
A multiple alignment using default settings was performed on the 12S sequences using the software package MUSCLE (Edgar 2004). A neighbor-joining tree for all sampled species was generated on MEGA11 (Kumar et al. 2018) using the Kimura-2-Parameter (K2P) model (Kimura 1980) with pairwise deletion and 1,000 bootstrap replicates for statistical support of the tree nodes. The rabbitfish, Chimaera monstrosa (NC_003136), was used as an outgroup to root the tree. Mean within- and between-group (intergeneric) distances with standard error estimates (1,000 bootstrap replications) were calculated using the same software and K2P model.
eDNA sample collection and extraction
Environmental DNA samples (n = 36) were collected from 13 locations (Fig. 2) during four expeditions between February 2020 and April 2021 (Online Resource 3). Two sets of samples (SW Cap St. Anne and Agadir) were collected during an expedition on an oceanographic vessel (R/V Amrigue). The 1 L Niskin bottles were released several times at each site for flushing before retrieving the final three sampling replicates. Samples were filtered using Sterivex™ Filter Units (Merck Millipore, 0.2 μm pore size) immediately upon collection on board the vessel. The remaining eDNA samples (n = 30) were obtained from inshore waters near villages or offshore waters within the PNBA using Polyethylene terephthalate (PET) plastic bottles during expeditions on traditional, local sailboats. Except for those collected at SW Cap St. Anne at 10–14 m depth, samples were collected just below the surface, covering habitats with different depths and vegetation profiles. These samples were filtered using Sterivex™ Filter Units (Merck Millipore, 0.2 or 0.45 μm pore size) within two to 48 h of sample collection. The final filtered volume for all sampling replicates was 750 mL. The filters were preserved either in Longmire buffer solution (0.1 M Tris, 0.1 M EDTA, 10 mM NaCl, 0.5% (w/v) SDS) (Longmire et al. 1997) or silica beads to prevent DNA degradation and stored at -20 °C until eDNA extraction in the lab. Each site was sampled in triplicate except for two sites with duplicates (Online Resource 3). DNA extractions were performed with the DNeasy® Blood & Tissue kit (Qiagen) following a modified eDNA extraction protocol from Spens et al. (2017). Extractions were electrophoresed on 0.8% agarose gel and quantified on Nanodrop 1000 (ThermoFisher). Extraction blanks and PCR blanks were performed and run with the remaining samples to check for possible contamination during the laboratory processing.
Library preparation and sequencing
Library preparation and sequencing were conducted at CIBIO. The MiFish-E (12S) primer set was used to amplify eDNA metabarcoding markers. All 36 samples, four extraction blanks, and three PCR blanks (one per batch of PCR replicates) were run on a parallel sequencing MiSeq platform (Illumina, San Diego, CA, USA), following a modified protocol from Miya et al. (2015) (Online Resource 4). PCR products from all sample replicates and blanks were cleaned with Ampure beads 0.8x, quantified in Nanodrop, pooled equimolar in a single library, and normalised to 15 nM. The library concentration was estimated using Nanodrop 1000 Spectrophotometer v3.8.1 (Thermo Fisher Scientific Inc.) and the quality was determined using Agilent TapeStation. A quantitative PCR (qPCR) was performed to validate and quantify the final library before sequencing on a single Illumina MiSeq run using the MiSeq v2 250PE kit at a concentration of 12 pM with 25% of PhiX.
Bioinformatic and statistical analysis
Analysis of demultiplexed raw reads was performed with the Anacapa Toolkit (Curd et al. 2019). A 12S genetic sequence reference database was tailored to this study with the Creating Reference libraries Using eXisting tools (CRUX) module. The database was created from newly generated barcodes for the species from this study and chondrichthyan reference sequences deposited in NCBI, excluding unverified entries and Chimaera species. Lastly, these barcodes were concatenated with a preexisting 12S database based on the MiFish-U primer set (Miya et al. 2015) provided by the Anacapa Toolkit. This step was included to avoid the incorrect assignment of amplified sequences. The final database consisted of 38,127 sequences of which 1,370 were assigned to Chondrichthyes, amounting to approximately 396 elasmobranch species. Lastly, the Quality Control and ASV parsing modules were run on default settings with the custom-made 12S reference database following the protocol used by Curd et al. (2019).
Only class Chondrichthyes was considered for analysis. To limit the probability of including false positive results from potential contamination, species represented by a single sequence read across all 13 sample locations were excluded from the analysis. All taxonomic assignments below species level were excluded except for ASVs assigned to the genus Myliobatis, since a single species in this genus is described from the East Atlantic (the common eagle ray: Myliobatis aquila), which has previously been recorded in PNBA landings. Furthermore, species with distribution ranges that did not include the eastern Atlantic were assumed to be erroneously assigned or to be the product of errors in amplification, sequencing, or reference databases and were removed from the final dataset. Five species had high read counts (≥ 20 reads) across two extraction blanks, suggesting some level of contamination during the DNA extraction process of some samples. In the complete dataset, over 90% of reads for the whipray complex, Fontitrygon margarita/margaritella, and the blue shark, Prionace glauca, stemmed from extraction blanks. In comparison, 55%, 18%, and 0.6% of reads for the spiny dogfish, Squalus acanthias, the common guitarfish, Rhinobatos rhinobatos, and the milk shark, Rhizoprionodon acutus, were found in extraction blanks, respectively. To avoid including potential false positive species detections and based on the lack of physical records of the species in the PNBA, Prionace glauca and S. acanthias were excluded from all analyses. The other three species were excluded from the environmental samples corresponding to the contaminated extraction blanks. Still, they were retained in other samples as accurate detections given their consistent reports within the PNBA before (e.g., Barham et al. 2011) and during the study period. PCR replicates (n = 3 per sample) with no or only a single read were discarded and reads from the remaining PCR replicates and sample replicates were summed up into a single unit per site (Online Resource 5).
Taxon diversity, community composition, and read abundance were explored through α- and β- biodiversity indices using presence/absence data and read abundance data. Community composition across sites was described using species richness (S) and Shannon’s (H) diversity index to explore possible differences within northern, central, and southern, or near- and offshore communities within the PNBA. Differences between samples were assessed for significance using a Kruskal-Wallis test for non-parametric datasets. Sample-based species accumulation curves were created to evaluate the completeness of sampling. All analyses were conducted using the vegan v2.6-2 package on R Studio v2022.7.1.554 (RStudio Team 2022).
PNBA species list
To monitor the integrity of the reference database created for the PNBA, a custom list of elasmobranch species either recorded or suspected to occur in the PNBA was compiled (Online Resource 6) using species identification books (Last et al. 2016; Ebert et al. 2021), records from online open-access databases such as the Global Biodiversity Information Facility (GBIF, www.gbif.org) and the Ocean Biogeographic Information System (OBIS, www.obis.org), peer-reviewed and grey literature (Jager 1993; Ducrocq et al. 2004; Valadou et al. 2006; Diop and Dossa 2011; Séret and Naylor 2016; Araujo and Campredon 2018; Lemrabott 2023), and reports from the Institut Mauritanien de Recherches Océanographiques et de Pêches (IMROP) (Barham et al. 2011).
Considering the shallow nature of the Banc d’Arguin (0–30 m), only species with recorded distribution in Mauritania, habitat preferences encompassing depths between 0 and 50 m, or whose presence was confirmed through personal observations in the PNBA, were included.
Results
DNA barcoding and species identification
A total of 217 tissue samples were collected and initially identified to the lowest taxonomic category possible based on morphological features (Online Resource 1). Although morphology was instrumental in confirming the assignment of species to genus or family levels, species-level identification was compromised in several cases due to specimens having undergone processing at the time of sampling, i.e., not presenting recognizable features such as dorsal or pectoral fins or coloration patterns due to drying.
All samples were amplified for 12S (sequence lengths between 145 and 183 bp); however, 14 sequences were discarded due to poor quality. None of the sequences that were discarded belonged to putative species for which only a single specimen was sampled; thus, it was not expected to have affected the species’ representation in the final dataset. The morphological identification of 23 specimens was corrected at species and genus levels based on molecular identification. The final 12S dataset included 203 ASVs assigned to 26 confirmed species and two putative new species from 24 different genera, 16 families, and eight orders (Fig. 3). Twelve shark species from 11 genera, nine families and four orders and 16 ray species from 13 genera, seven families and four orders were present. Of 39 and 10 samples amplified for COI and NADH2, respectively, 32 COI sequences (606–655 bp) and five NADH2 sequences (824–1339 bp) had sufficient quality for molecular identification. Most taxa with available references were readily identified based on 12S alone (Table 1), with some exceptions presented below.
Neighbour-joining tree for shark and ray species sampled in Mauritania based on 12S barcodes, using Kimura-2-Parameter Model and 1,000 bootstrap replicates. Bootstrap values above 50% are displayed. Species are coloured according to taxonomic order to display lineage diversity within the region. Chimaera monstrosa (NC_003136) is used as an outgroup to root the tree. Numbers in brackets indicate the number of specimens included in the construction of the tree. Taxa not resolved to species level indicated by genus (e.g., Torpedo)
Sharks
The dusky shark, Carcharhinus obscurus, could not be confidently distinguished from its congener C. galapagensis with either marker, as 12S, COI and NADH2 sequences all showed 100% pairwise similarity with both species. Both species occur in Northwest Africa and can be challenging to distinguish morphologically and genetically at the mitochondrial level (Corrigan et al. 2017). In the East Pacific, they are known to hybridise (Pazmiño et al. 2019), making genetic identification more challenging using only mitochondrial markers. Here, we consider only C. obscurus since C. galapagensis has not previously been reported from the PNBA. However, we note that further research is required on this topic.
Based on recognized distribution ranges, the blackspotted smoothhound, Mustelus punctulatus, was first presumed to be the common smoothhound, Mustelus mustelus. Mustelus punctulatus is known to occur in the Mediterranean and East Atlantic coastal regions north of Western Sahara, while the presence of M. mustelus in the study area is well established in the literature. However, none of the 12S sequences generated from sampled specimens matched M. mustelus (92.3%) or produce any species matches above 95% similarity (M. manazo 94.9%) with BLAST, likely due to low taxon coverage (six out of 27 species, M. punctulatus not included). Mean interspecific genetic distance for 12S is 4.22 ± 1.01%. In turn, NADH2 coverage for the genus Mustelus includes 22 species, and a single amplified NADH2 sequence confirmed the species as M. punctulatus (99.7%) (Fig. 4d).
Rays
Four specimens identified as Gymnura based on morphology and genetic results had no taxon matches above 98% similarity for 12S and COI (NADH2 amplification was unsuccessful). The closest match on GenBank was the spiny butterfly ray, G. altavela, with 93.9% and 91.5% similarity for 12S and COI, respectively. While taxon coverage for Gymnura spp. is low for 12S (4 out of 12 described species), all known species had COI reference sequences except for two (G. tentaculata, described from the Indo-West Pacific, and G. sereti, described from the Central East Atlantic, including Mauritania). However, the unidentified Gymnura species can be distinguished from G. sereti (Yokota and Carvalho 2017) based on conspicuous physical characters, i.e., the presence of spiracular tentacles and a distinctly long tail (± 45% of disc width, visual estimation). Mean interspecific distance for 12S is 7.75 ± 1.36% and 15.3 ± 1.17% for COI. Thus, evidence from morphological characteristics, genetics, and geographical distribution, suggests that the species is potentially new to science (Fig. 4a) and is referred to as “Gymnura sp.”.
One Torpedo specimen caught during a beam trawl survey in the PNBA could not be morphologically identified at the species level (Fig. 3b). While COI and NADH2 amplification was unsuccessful, 12S matched most closely with T. marmorata (93.3%). Four out of ten known Torpedo species fall into the studied geographical range (T. bauchotae, T. mackayana, T. marmorata, T. torpedo; Last et al. 2016), but global taxon coverage for the genus and 12S marker includes only three species (T. marmorata, T. tokionis, and T. sinuspersici). Average interspecific pairwise distances for 12S of these species (n = 4 including unidentified species from this study) is 6.37 ± 1.41%. Coloration patterns of other described species do not match our specimen. All evidence indicates it is likely an undescribed species, henceforth referred to as “Torpedo sp.” (Fig. 4b).
Specimens identified as the Lusitanian cownose ray, Rhinoptera marginata had 100% similarity with R. brasiliensis and a close match with R. javanica (99.4%); however, R. marginata had no 12S reference sequences available. COI sequences from these specimens were a 99.9% match with R. marginata, and between 98.9 and 99.4% match for R. brasiliensis. Three and six out of eight known Rhinoptera species had available reference sequences for 12S and COI, with mean interspecific genetic distances for 12S and COI of 2.2 ± 0.71% and 5.02 ± 0.61%, respectively. The Rhinoptera genus is relatively poorly known and taxonomic placement based on genetics and morphology are often challenging (Naylor et al. 2012), however, the species was tentatively kept as R. marginata due to its currently accepted geographical distribution (East Atlantic) compared to R. brasiliensis (West Atlantic) and R. javanica (Indo-West Pacific) (Last et al. 2016).
The duckbill eagle ray, Aetomylaeus bovinus, formed two distinct clusters each represented by a minimum of three specimens (Fig. 3), with an intra-specific genetic distance of 0.49 ± 0.34%. The other two markers failed to amplify. We did not detect noticeable morphological differences between the sampled specimens, and all were identified as A. bovinus.
One taxonomic group that remains contested is the genus Fontitrygon. Differences between the daisy whipray, F. margarita, and the pearl whipray, F. margaritella, could not confidently be resolved based on morphological characteristics or DNA barcoding of 12S and COI markers (NADH2 amplification was unsuccessful). These specimens were thus identified as part of the F. margarita/margaritella species complex.
Elasmobranch species diversity in eDNA
A total of 2,657,521 raw reads were extracted from 36 physical samples as well as two PCR blanks and four extraction blanks. After completing quality control protocols and assigning taxonomy to filtered sequence reads, 1,709,284 reads were retained and appointed to one of 429,183 distinct ASVs. Taxa from six eukaryotic taxonomic groups, including Actinopterygii, Amphibian, Aves, Chondrichthyes, Mammalia, and Petromyzontida were identified, with the highest number of reads assigned to Actinopterygii (28.21%; n = 482,167). However, only elasmobranchs were considered for this study and accounted for 7.44% of total filtered reads (n = 127,249). After removing potential contamination errors, singletons, and ASVs presumed to be erroneously assigned to geographically improbable species, a total of 27 species were identified. These included 10 species of sharks and 17 species of rays, including an unidentified Myliobatis species (Fig. 2a, b). Elasmobranch taxa were recovered in 100% of the samples, with samples collectively displaying an average species richness (S) of 15 (range 9–19). The most diverse sites were West Tidra 1 and 2 (S = 19 and 18, respectively) as well as L’oeil (S = 18).
Two species were present across all samples, namely G. altavela and the striped panray, Zanobatus schoenleinii, however, species composition varied across all samples with no distinct patterns emerging across northern, central, and southern regions or between near-shore or offshore sample sites (Online Resource 7).
Based on habitat preferences and depth distribution of regionally recorded species, 57 species were found to potentially use the area (Online Resource 6). Previous catch records from the PNBA have documented the presence of at least 31 elasmobranch species (counting Raja sp. as one taxonomic unit as noted in reports), although many may presently be very rare or regionally extinct. Of these species, 15 were detected in eDNA samples, while 12 species are new (official) records, such as the lesser spotted dogfish, Scyliorhinus canicula, the smalltooth stingray Hypanus rudis (Fig. 4c), and the pelagic stingray, Pteroplatytrygon violacea. These were retained as true positives since S. canicula had previously been recorded (Jabado, unpubl. data) and the latter two had been observed either at the Nouadhibou processing site or in the PNBA (author, pers. observation). Large pelagic sharks such as the common thresher, Alopias vulpinus, and the shortfin mako, Isurus oxyrinchus, as well as species associated with deeper habitats such as the sharpnose sevengill shark, Heptranchias perlo, had not been previously reported but were retained as true positives due to their widespread and highly migratory nature. One of the most common species in the region, M. mustelus, was not detected. Instead, M. punctulatus was recorded in 77% of samples. Sphyrna lewini and the smooth hammerhead, S. zygaena, were not detected in eDNA samples, although their presence in the PNBA is well recorded. Six previously reported species (whitespotted guitarfish, Rhinobatos albomaculatus, spineback guitarfish, R. irvinei, African wedgefish, Rhynchobatus luebberti, night shark, Carcharhinus signatus, Séret’s butterfly ray, Gymnura sereti, and Negaprion brevirostris) were missing a 12S reference sequence at the time of analysis and could therefore not be detected (Fig. 6).
Species accumulation curves are plotted to show elasmobranch diversity as a function of the number of eDNA samples taken inside the PNBA (Fig. 5). The curve flattens after approximately eight samples when considering all species combined, suggesting that a higher sampling effort would likely not significantly increase the observed diversity.
Conservation status
Of 27 species positively identified from samples taken inside the PNBA boundaries, seven species are Critically Endangered (25%), assuming the presence of M. aquila and including S. lewini, which despite not being detected in eDNA samples was visually confirmed in the PNBA during our sampling period. Six species are Endangered (22.2%) and six, or seven if including F. margarita, are Vulnerable (22.2% or 25.9%) according to the IUCN Red List (Fig. 6; IUCN 2022). Species assessed as threatened with extinction in the PNBA therefore amount to 67.9% (including S. lewini). Of the remaining species, four are Near Threatened (14.8%), or five if including F. margaritella (18.5%), two are Least Concern (7.4%), and two putative new species from this study have not been evaluated.
Species reported in the literature to occur in the PNBA (left) compared to species detected in eDNA samples not previously reported from the area (right), as well as species both previously reported and detected in eDNA samples (intersection). Species in bold were verified visually during or after sampling period. Asterisks denote species which are known to have occurred in the PNBA in the past but with no published landing records a, species for which 12S reference sequences were not available and could not be obtained b, or species for which 12S reference sequences were obtained from private databases (Séret, B., and Naylor, G., unpublished data) c. Species are categorised according to their IUCN Red List status
Discussion
This study provides the first eDNA survey effort in Mauritania and a first exhaustive regional barcoding effort for elasmobranchs in West Africa. Results illustrate the importance of molecular tools for uncovering overlooked and cryptic diversity. We provide a database with new barcodes for almost half the species detected, which proved instrumental in producing species-specific identifications from metabarcoding. We also provide the first consolidated and fisheries-independent species checklist for the PNBA, improving previous knowledge derived from catch and landing reports (e.g., Ducrocq et al. 2004; Barham et al. 2011).
Species diversity and taxonomic changes
Relatively high species diversity is reported from the Banc d’Arguin, with at least 27 elasmobranch species detected in eDNA samples. This is more than double the number of species detected through a similar metabarcoding study conducted in the Bijagós Islands (Guinea-Bissau) with a higher sampling effort (Leurs et al. 2023). Landings surveys in The Gambia and Ghana detected 27 and 34 species, respectively, with more extensive sampling and monitoring periods (Moore et al. 2019; Seidu et al. 2022a). When adding recent, verified IMROP observer records, pictures collected from local fishers, and personal observations during the sampling period, at least three additional species can be added to the PNBA species checklist (Sphyrna lewini, Galeocerdo cuvier, and Carcharhinus leucas), increasing the number of species to 30. Addressing data deficiencies on species diversity in shallow coastal habitats is important, as the Banc d’Arguin and the Bijagós Islands are both considered key sites for elasmobranchs in West Africa (Ducrocq et al. 2004; Diop and Dossa 2011).
Most species encountered were expected to be found in the PNBA based on local fisheries catch and landings data (Jager 1993; Barham et al. 2011; Trégarot et al. 2020) or the known species range (Last et al. 2016; Ebert et al. 2021). However, some exceptions exemplify the potential consequences of taxonomic uncertainty and misidentifications to species conservation. Notably, M. punctulatus was detected instead of M. mustelus in eDNA samples. Similarly, Mustelus tissue samples collected during two different seasons and sampling events were all identified as M. punctulatus, although M. mustelus is reportedly commonly captured in small-scale and industrial fisheries (Ducrocq et al. 2004; Failler et al. 2006; Gascuel et al. 2007; Barham et al. 2011). In Mauritania, M. mustelus reportedly moves to coastal areas following decreasing sea temperatures during the cold season from January to May (Khallahi 2004). Yet, samples (i.e., tissue and eDNA samples) taken during and outside this season only detected M. punctulatus. Similarly, Guardone et al. (2017) barcoded different species imported into Europe and found that species labelled as M. mustelus originating from Mauritania were, in fact, M. punctulatus. This raises the urgent question of whether M. punctulatus, a species that is morphologically similar and can be mistaken for M. mustelus (Marino et al. 2018; Ebert et al. 2021), is in fact predominant in the region and regularly misidentified, or whether both species co-occur with other factors (e.g., sample size, sampling area) driving the absence of M. mustelus from our samples. Overall, findings suggest that M. punctulatus may have a wider distribution in the Central-East Atlantic than previously documented and that the distribution range of M. mustelus along the western coast of Africa, where population declines of up to 80% have been noted (Jabado et al. 2021a), may be narrower or more fragmented. This could increase the conservation threat to M. punctulatus if connectivity between isolated populations is limited and fishing pressure is high (Boussarie et al. 2022). Considering that M. mustelus (“tollo”) is one of two shark species that can legally be targeted in the PNBA (FAO 2018), research needs to be done to delimit Mustelus populations to determine the sustainability of this fishery, especially as both species are threatened and Mustelus species are some of the most landed and traded sharks in Mauritania (Jabado, unpubl. data).
Some misidentifications were also noted in the Dasyatidae family, where Hypanus rudis represents a new species record for the PNBA and Mauritania. The species is frequent in PNBA landings but may regularly be confused with its congeners (Jabado et al. 2021b; author pers. observation). However, stingrays (and skates) are also often reported in aggregated taxonomic categories (i.e., Dasyatidae, Raja spp.), likely masking the true species diversity within the group (Failler et al. 2006; Barham et al. 2011). This is exemplified by the absence of other dasyatid rays like P. violacea or T. grabatus in published records, or the absence of any species-specific record of skates within the PNBA (Barham et al. 2011). However, access to databases from the IMROP is restricted and data collected are often not accessible. Hypanus rudis has been assessed as Critically Endangered, but its occurrence in the region warrants further research to improve understanding of its actual distribution, which is presently inferred from a few records between Cameroon and Senegal (Moore et al. 2019; Petean et al. 2020; Jabado et al. 2021b; Leurs et al. 2023).
Changes in reported species diversity also stem from the discovery of potentially new species. Gymnura species reported and traded as G. altavela in Mauritania, could in fact be two separate species. Cryptic speciation of G. altavela was also reported recently from both sides of the Atlantic and the Mediterranean Sea, further supporting our results (Vilasboa et al. 2022; Cady et al. 2023). Gymnura species are some of the most abundantly traded rays in Mauritania (Jabado, unpubl. data), and population declines of 42.5 and 54% have been estimated for Morocco and Senegal, respectively, over the last few decades (Dulvy et al. 2021b). This raises concerns about the sustainability of their exploitation in the PNBA, especially as the species may in fact represent two separate management units. While the actual regional abundance of G. altavela may have been overestimated, the distribution of Gymnura sp. may extend beyond Mauritania but would currently not be recorded, warranting further genetic and morphological research to determine the species distribution and abundance along its range. Within the PNBA, where Gymnura spp. are reportedly incidentally captured, over 90% of individuals are juveniles or subadults (Dulvy et al. 2021b), indicating the need to implement bycatch reduction measures. While Torpedo sp. is also proposed as a putative new species, electric rays are not usually retained and are discarded in most local fisheries (Imraguen fisher, pers. comm.). No voucher specimens could be retained (but photos were taken) for specimens of Gymnura and Torpedo, and 12S databases lack sufficient species representation within their respective genera. Therefore, further biometric data collection on both species is essential to confirm current findings.
Overall, evidence suggests that management of several taxa is currently based on erroneous, inaccurate, or incomplete species data. This stresses the need to invest in capacity building and training of local fisheries observers to improve the quality and accuracy of collected data in field locations where other types of monitoring may be financially or logistically restricted.
Extinctions and declines of large-bodied elasmobranch species
Results also support accounts of the regional or local extinction or near-extinction of some large-bodied shark and ray species, including sawfishes (Pristis spp.) (Robillard and Séret 2006), wedgefishes (Rhynchobatus spp.) (Kyne et al. 2020; Jabado, unpubl. data), lemon sharks (Negaprion brevirostris) (Diop and Dossa 2011), tiger sharks (Galeocerdo cuvier) (Araujo and Campredon 2018), and the recently described endemic false shark ray (Rhynchorhina mauritaniensis) (Séret and Naylor 2016). These were not sighted at processing sites (except one tiger shark) or detected in eDNA samples, neither at species nor genus levels. Although some of these species lacked a corresponding reference sequence, potentially leading to false negative results, ASVs of such species (if present) would be expected to be assigned to genus level represented by other congeners in the database (e.g., Pristis, Rhynchobatus, Negaprion). Their apparent absence is consistent with similar reports from other West African countries, such as Ghana (Seidu et al. 2022b), Senegal, The Gambia, and Guinea-Bissau (Diop and Dossa 2011), where these species have become increasingly rare or locally extinct. Factors such as collected water volume, sampling depth, strong tidal fluctuations, and seasonality (most samples were collected just below the sea surface between November and April) may have influenced detection rates of rare, bottom-dwelling species, or migratory species (Hansen et al. 2018; Bessey et al. 2020). Nevertheless, informal reports from local fishers also point towards the absence of these (reportedly) once abundant species in local waters.
The few larger shark species that still occur in the PNBA such as C. obscurus and S. lewini, may also be at risk, as catches throughout the region are becoming rarer (Diop and Dossa 2011; Lemrabott 2023). Although the ecological ramifications of these species losses and declines within the Banc d’Arguin have not been explored, Lemrabott (2023) suggested that the consistent overfishing and subsequent decline of larger shark species, such as S. lewini, could have instigated a trophic cascade affecting the abundance of their prey (e.g., R. marginata) and bivalve species consumed by the latter. The accelerated pace of local and regional population declines and species extinctions, and their potential ecological consequences warrants further research and the development of more efficient conservation strategies.
Conservation status and threats
While globally an estimated one third of all chondrichthyan species are considered threatened (Critically Endangered, Endangered, or Vulnerable) according to the IUCN Red List of Threatened Species (Dulvy et al. 2021a), over two thirds (67.9%) of elasmobranch species detected in eDNA samples in the PNBA are threatened with extinction (including S. lewini). These numbers are alarming considering that overall elasmobranch landings do not appear to have decreased since the introduction of the elasmobranch-specific fishing ban in 2003 (Westlund et al. 2017; Trégarot et al. 2020). Further, these landings mostly comprise threatened species (e.g., G. cemiculus, R. marginata, R. acutus, A. bovinus, S. lewini, and G. cirratum) (Barham et al. 2011; Lemrabott 2023). Ongoing exploitation of these species in the PNBA is likely unsustainable and immediate action is required to revise current management strategies. This is reflected in the local disappearance of larger predatory sharks (Diop and Dossa 2011) and the long-term change in size structure in heavily exploited species such as S. lewini and G. cemiculus (Walker et al. 2005; Barham et al. 2011; Lemrabott 2023). Furthermore, the Banc d’Arguin is supposedly a reproduction area for several elasmobranch species (Valadou et al. 2006), including the Critically Endangered S. lewini (de la Hoz Schilling, unpubl. data). and many species detected in this survey are highly mobile and likely regularly move outside PNBA boundaries, where industrial fisheries occupy large parts of the PNBA buffer zone, increasing fishing pressure on them (Leurs et al. 2021).
eDNA-based monitoring in remote and isolated field locations
Despite challenging field conditions, this study established the usefulness of eDNA in detecting elasmobranch species in remote and relatively unexplored locations such as the Banc d’Arguin, with high confidence levels. Every species barcoded individually was retrieved in at least one eDNA sample, except for G. cuvier and Alopias superciliosus, although A. vulpinus was detected. Records of certain pelagic and highly mobile species not previously reported from the PNBA (e.g., A. vulpinus, I. oxyrinchus, H. perlo) suggest that these species may not be common there but may use its waters opportunistically. However, some species whose presence in the area is well established and verified, such as Sphyrna lewini and S. zygaena, were not detected in eDNA samples. This could be due to a seasonal and/or spatial factor, as S. lewini, for example, is thought to use the Banc d’Arguin as a pupping ground between April–July at sites not sampled during this study (de la Hoz Schilling, unpubl. data). Although eDNA has been proven to be a useful tool to record species richness and composition in remote locations that are difficult to monitor, results often do not concur with visual surveys (Boussarie et al. 2018; Dunn et al. 2023; Leurs et al. 2023). Nevertheless, most species reported from catches in the PNBA in the last decade (considering likely misidentifications) were detected in eDNA samples (Lemrabott 2023) and species accumulation curves indicate that an increased sampling effort would not yield significantly more species diversity. Although this demonstrates the efficiency of metabarcoding as a complementary monitoring tool, some adjustments to the sampling design (i.e., sampled area, seasonality, sampling depth, filtered sample volume) could maximise the number of taxa detected (Bessey et al. 2020).
Recommendations for conservation actions and priorities
This assessment of current regional diversity is a baseline for future research on the distribution and abundance of sharks and rays within the PNBA and Mauritania. However, conservation concerns discussed should be addressed swiftly, requiring immediate action to mitigate further population declines and local species extinctions, especially as most detected species are of global conservation concern, and lack data to assess threat levels to local populations. Current management plans (i.e., blanket bans on all elasmobranch species except for M. mustelus and L. smithii) and laws protecting elasmobranchs in the PNBA (Loi n° 2000.024) by allowing exclusively subsistence fishing (defined by Loi N°2000-025 du Code des Pêches de Mauritanie) are insufficiently enforced and have proven inefficient at significantly reducing elasmobranch landings (FAO 2018), although these are not consumed locally (Jabado, unpubl. data). While local capacity building and bycatch reduction measures (e.g., training fishers on safe handling and release, reduced gillnet soak times) may be useful in aiding elasmobranch conservation, the profitability of elasmobranch fisheries and the socio-economic susceptibility of the Imraguen fisher community are factors likely impeding compliance with existing regulations (Westlund et al. 2017). Indeed, blanket bans on elasmobranch fishing often encounter local resistance, leading to the development of illegal activities (e.g., Diop and Dossa 2011; Carr et al. 2013; Vianna et al. 2016; Trégarot et al. 2020). Therefore, and in addition to research gaps discussed above, viable options for elasmobranch conservation in the PNBA may include spatio-temporal closures based on the identification of species-specific reproductive areas, incentivizing the release of juvenile or immature specimens (Booth et al. 2023), supporting alternative livelihoods options, and enforcing existing policies.
We provide a first list of elasmobranch species in the PNBA confirmed through multiple approaches, however, more work is necessary. Considering the dire conservation status of most elasmobranch species occurring in the PNBA and the continuing fishing pressure they are exposed to in and outside this area, current monitoring and regulatory strategies require immediate enforcement.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Araujo A, Campredon P (2018) Banc d’Arguin (Mauritania). In: Finlayson CM, Milton GR, Prentice RC, Davidson NC (eds) The Wetland Book. II: distribution, description and conservation. Springer, Dordrecht, pp 1319–1332. https://doi.org/10.1007/978-94-007-4001-3
Barham CB, Yerba L, Sakho CI, Ahmed Deida H (2011) Évolution de l’activité de pêche en 2009 au PNBA: effets environnementaux, rôles écologiques et socio-économiques. Rapport groupe de travail PSPI. IMROP, Iwik, Mauritania, 15–17 juin 2010. http://hdl.handle.net/1834/4665
Bessey C, Jarman SN, Berry O, Olsen YS, Bunce M, Simpson T, Power M, McLaughlin J, Edgar GJ, Keesing J (2020) Maximizing fish detection with eDNA metabarcoding. Environ DNA 2(4):493–504. https://doi.org/10.1002/edn3.74
Booth H, Ramdlan MS, Hafizh A, Wongsopatty K, Mourato S, Pienkowski T, Adrinato L, Milner-Gulland EJ (2023) Designing locally-appropriate conservation incentives for small-scale fishers. Biol Conserv 277:109821. https://doi.org/10.1016/j.biocon.2022.109821
Boulay S (2013) Pêcheurs Imraguen Du Sahara atlantique: mutations techniques et changements sociaux des années 1970 à nos jours. Karthala, Paris
Boussarie G, Bakker J, Wangensteen OS, Mariani S, Bonnin L, Juhel JB, Kiszka JJ, Kulbicki M, Manel S, Robbins WD, Vigliola L, Mouillot D (2018) Environmental DNA illuminates the dark diversity of sharks. Sci Adv 4:eaap9661. https://doi.org/10.1126/sciadv.aap9661
Boussarie G, Momigliano P, Robbins WD, Bonnin L, Cornu JF, Fauvelot C, Kiszka JJ, Manel S, Mouillot D, Vigliola L (2022) Identifying barriers to gene flow and hierarchical conservation units from seascape genomics: a modelling framework applied to a marine predator. https://doi.org/10.1111/ecog.06158. Ecography e06158
Budd AM, Cooper MK, Schils T, Mills MS, Deinhart ME, Huerlimann R, Strugnell JM (2021) First detection of critically endangered scalloped hammerhead sharks (Sphyrna lewini) in Guam, Micronesia, in five decades using environmental DNA. Ecol Indic 127:107649. https://doi.org/10.1016/j.ecolind.2021.107649
Burgess GH, Beerkircher LR, Cailliet GM, Carlson JK, Cortés E, Goldman KJ, Grubbs RD, Musick JA, Musyl MK, Simpfendorfer CA (2005) Is the collapse of shark populations in the Northwest Atlantic Ocean and Gulf of. Mexico real? Fish 30:19–26. https://doi.org/10.1577/1548-8446(2005)30[19:ITCOSP]2.0.CO;2
Cady T, Bemis KE, Baeza JA (2023) The mitochondrial genome of the endangered Spiny Butterfly Ray Gymnura altavela (Linnaeus 1758) (Myliobatiformes: Gymnuridae) provides insights into cryptic lineages. Mitochondrial DNA Part A. https://doi.org/10.1080/24701394.2023.2251577
Carr LA, Stier AC, Fietz K, Montero I, Gallagher AJ, Bruno JF (2013) Illegal shark fishing in the Galápagos Marine Reserve. Mar Policy 39:317–321. https://doi.org/10.1016/j.marpol.2012.12.005
Cashion MS, Bailly N, Pauly D (2019) Official catch data underrepresent shark and ray taxa caught in Mediterranean and Black Sea fisheries. Mar Policy 105:1–9. https://doi.org/10.1016/j.marpol.2019.02.041
Collins RA, Bakker J, Wangensteen OS, Soto AZ, Corrigan L, Sims DW, Genner MJ, Mariani S (2019) Non-specific amplification compromises environmental DNA metabarcoding with COI. Methods Ecol Evol 10:1985–2001. https://doi.org/10.1111/2041-210X.13276
Cornejo R, Kouri JC, Vélez-Zuazo X, González-Pestana A, Mucientes G (2015) An updated checklist of Chondrichthyes from the Southeast Pacific off Peru. Check List 11(6):1809. https://doi.org/10.15560/11.6.1809
Corrigan S, Delser PM, Eddy C, Duffy C, Yang L, Li C, Bazinet AL, Mona S, Naylor GJP (2017) Historical introgression drives pervasive mitochondrial admixture between two species of pelagic sharks. Mol Phylogenet Evol 110:122–126. https://doi.org/10.1016/j.ympev.2017.03.011
Curd EE, Gold Z, Kandlikar GS, Gomer J, Ogden M, O’connell T, Pipes L, Schweizer TM, Rabichow L, Lin M, Shi B, Barber PH, Kraft N, Wayne R, Meyer RS (2019) Anacapa Toolkit: an environmental DNA toolkit for processing multilocus metabarcode datasets. Methods Ecol Evol 10:1469–1475. https://doi.org/10.1111/2041-210x.13214
Dent F, Clarke S (2015) State of the global market for shark products. FAO Fisheries and Aquaculture Technical Paper N° 590. FAO, Rome
Diop M, Dossa J (2011) 30 Years of shark fishing in West Africa: development of fisheries, catch trends, and their conservation status in Sub-Regional Fishing Commission member countries. Report to FIBA. https://www.iucnssg.org/uploads/5/4/1/2/54120303/30years_eng.pdf
Ducrocq M, Lemine Ould Sidi M, Ould Yarba L (2004) Comment le Parc National du Banc d’Arguin est devenu le plus grand sanctuaire d’Afrique pour les requins. Report to PRCM. https://portals.iucn.org/library/sites/library/files/styles/publication/public/book_covers/BC-2004-133.jpg
Dulvy NK, Pacoureau N, Rigby CL et al (2021a) Overfishing drives over one-third of all sharks and rays toward a global extinction crisis. Curr Biol 31:1–15. https://doi.org/10.1016/j.cub.2021.08.062
Dulvy NK, Charvet P, Carlson J et al (2021b) Gymnura altavela The IUCN Red List of Threatened Species e. https://doi.org/10.2305/IUCN.UK.2021-1.RLTS.T63153A3123409.en. .T63153A3123409
Dunn N, Curnick DJ, Carbone C, Carlisle AB, Chapple TK, Dowell R, Ferretti F, Jacoby DMP, Schallert RJ, Steyaert M, Tickler DM, Williamson MJ, Block BA, Savolainen V (2023) Environmental DNA helps reveal reef shark distribution across a remote archipelago. Ecol Indic 154:110718. https://doi.org/10.1016/j.ecolind.2023.110718
Ebert DA, Dando M, Fowler S (2021) Sharks of the World: a complete guide. Princeton University Press, New Jersey
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Erhardt T, Weder R (2020) Shark hunting: on the vulnerability of resources with heterogeneous species. Resour Energy Econ 61:101181. https://doi.org/10.1016/j.reseneeco.2020.101181
Failler P, Diop M, Dia MA, O/Inejih CA, Tous P (2006) Evaluation des stocks et aménagement des pêcheries de la ZEE mauritanienne. Rapport du cinquième Groupe de travail IMROP. Nouadhibou, Mauritanie, 9–17 décembre 2002. COPACE/PACE Séries No. 05/66, FAO, Rome
Failler P, Deme M, Diop A, Balbé D, da Silva A, Daim Dia A, Bakalakiba A (2009) Les aires protégées estuariennes, côtières et marines (APECM) en afrique de l’Ouest: des réservoirs de ressources aquatiques en sursis. Revue Africaine Des Affaires Maritimes et des Transports 1:44–49
FAO (2014) The state of World fisheries and Aquaculture: opportunities and challenges. FAO, Rome
FAO (2018) A country and regional prioritisation for supporting implementation of CITES provisions for sharks. Fisheries and Aquaculture Circular No. 1156, FAO, Rome
FAO (2023) CECAF (Eastern Central Atlantic) capture production. Fisheries and Aquaculture Division [online], Rome
Fields AT, Fischer GA, Shea SK, Zhang H, Abercrombie DL, Feldheim KA, Babcock EA, Chapman DD (2018) Species composition of the international shark fin trade assessed through a retail-market survey in Hong Kong. Conserv Biol 32:376–389. https://doi.org/10.1111/cobi.13043
Fricke R, Mahafina J, Behivoke F, Jaonalison H, Léopold M, Ponton D (2018) Annotated checklist of the fishes of Madagascar, southwestern Indian Ocean, with 158 new records. FishTaxa 3(1):1–432
Gascuel D, Labrosse P, Meissa B, Taleb Sidi MO, Guénette S (2007) Decline of demersal resources in North-West Africa: an analysis of Mauritanian trawl-survey data over the past 25 years. Afr J Mar Sci 29:331–345. https://doi.org/10.2989/AJMS.2007.29.3.3.333
Guardone L, Tinacci L, Costanzo F, Azzarelli D, D’Amico P, Tasselli G, Magni A, Guidi A, Nucera D, Armani A (2017) DNA barcoding as a tool for detecting mislabeling of fishery products imported from third countries: an official survey conducted at the Border Inspection Post of Livorno-Pisa (Italy). Food Control 80:204–216. https://doi.org/10.1016/j.foodcont.2017.03.056
Hansen BK, Bekkevold D, Clausen LW, Nielsen EE (2018) The sceptical optimist: challenges and perspectives for the application of environmental DNA in marine fisheries. Fish Fish 19:751–768. https://doi.org/10.1111/faf.12286
Hebert PDN, Cywinska A, Ball SL, Jeremy R (2003a) Biological identifications through DNA barcodes. Proc Biol Sci 270:313–321. https://doi.org/10.1098/rspb.2002.2218
Hebert PDN, Ratnasingham S, DeWaard JR (2003b) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci 270:S96–S99. https://doi.org/10.1098/rsbl.2003.0025
IUCN (2022) The IUCN Red list of threatened species. Version 2021-2. https://www.iucnredlist.org. Downloaded on 15 Sep 2022.
Jabado RW, Chartrain E, Cliff G et al (2021a) Mustelus mustelus. The IUCN Red List of Threatened Species 2021: e.T39358A124405881. https://doi.org/10.2305/IUCN.UK.2021-2.RLTS.T39358A124405881.en
Jabado RW, De Bruyne G, Derrick D, Doherty P, Diop M, Leurs GHL, Metcalfe K, Porriños G, Seidu I, Tamo A, VanderWright WJ, Williams AB (2021b) Hypanus rudis. The IUCN Red List of Threatened Species 2021: e.T161620A124516434. https://doi.org/10.2305/IUCN.UK.2021-2.RLTS.T161620A124516434.en
Jager Z (1993) The distribution and abundance of young fish in the Banc d’Arguin, Mauritania. Hydrobiologia 258:185–196. https://doi.org/10.1007/BF00006196
Khallahi B (2004) Ecologie et biologie de l’émissole lisse Mustelus mustelus (Linné, 1758) sur les côtes de Mauritanie. Dissertation, Université de Bretagne Occidentale
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kyne PM, Jabado RW (2019) Glaucostegus cemiculus. The IUCN Red List of Threatened Species 2019: e.T104050689A104057239. https://doi.org/10.2305/IUCN.UK.2019-2.RLTS.T104050689A104057239.en
Kyne PM, Jabado RW, Rigby CL, Dharmadi Gore MA, Pollock CM, Herman KB, Cheok J, Ebert DA, Simpfendorfer CA, Dulvy NK (2020) The thin edge of the wedge: extremely high extinction risk in wedgefishes and giant guitarfishes. Aquat Conserv: Mar Freshw Ecosyst 30:1337–1361. https://doi.org/10.1002/aqc.3331
Lafferty KD, Garcia-Vedrenne AE, Mclaughlin JP, Childress JN, Morse MF, Jerde CL (2021) At Palmyra Atoll, the fish-community environmental DNA signal changes across habitats but not with tides. J Fish Biol 98:415–425. https://doi.org/10.1111/jfb.14403
Last PR, White WT, de Carvalho MR, Séret B, Stehmann MFW, Naylor GJP (2016) Rays of the World. CSIRO Publishing, Australia
Leeney RH, Downing N (2016) Sawfishes in the Gambia and Senegal - shifting baselines over 40 years. Aquat Conserv: Mar Freshw Ecosyst 26:265–278. https://doi.org/10.1002/aqc.2545
Lemrabott SY (2023) Fish and fisheries dynamics at Banc d’Arguin, Mauritania: Consequences for the intertidal food webs. Dissertation, University of Groningen. https://doi.org/10.33612/diss.574910952
Leurs G, van der Reijden KJ, Cheikhna Lemrabott SY, Barry I, Nonque DM, Olff H, Ledo Pontes S, Regalla A, Govers LL (2021) Industrial fishing Near West African Marine protected areas and its potential effects on Mobile Marine predators. Front Mar Sci 8:602917. https://doi.org/10.3389/fmars.2021.602917
Leurs G, Verkuil YI, Hijner N, Saalmann F, Dos Santos L, Regalla A, Ledo Pontes S, Yang L, Naylor GJP, Olff H, Govers LL (2023) Addressing data-deficiency of threatened sharks and rays in a highly dynamic coastal ecosystem using environmental DNA. Ecol Indic 154:110795. https://doi.org/10.1016/j.ecolind.2023.110795
Longmire JL, Maltbie M, Baker JB (1997) Use of lysis buffer in DNA isolation and its implication for museum collections. Museum of Texas Tech University 163:1–3
Mariani S, Jaquemet S, Baillie C, Magalon H (2021) Shark and ray diversity, abundance and temporal variation around an Indian Ocean Island, inferred by eDNA metabarcoding. Conserv Sci Pract 3:e407. https://doi.org/10.1111/csp2.407
Marino IAM, Finotto L, Colloca F, Di Lorenzo M, Gristina M, Farrell ED, Zane L, Mazzoldi C (2018) Resolving the ambiguities in the identification of two smooth-hound sharks (Mustelus mustelus and Mustelus punctulatus) using genetics and morphology. Mar Biodivers 48:1551–1562. https://doi.org/10.1007/s12526-017-0701-8
Marques V, Milhau T, Albouy C, Dejean T, Manel S, Mouillot D, Juhel JB (2021) GAPeDNA: assessing and mapping global species gaps in genetic databases for eDNA metabarcoding. Divers Distrib 27:1880–1892. https://doi.org/10.1111/ddi.13142
Miya M, Sato Y, Fukunaga T, Sado T, Poulsen JY, Sato K, Minamoto T, Yamamoto S, Yamanaka H, Araki H, Kondoh M, Iwasaki W (2015) MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R Soc Open Sci 2:150088. https://doi.org/10.1098/rsos.150088
Moore A, Séret B, Armstrong R (2019) Risks to biodiversity and coastal livelihoods from artisanal elasmobranch fisheries in a least developed country: the Gambia (West Africa). Biodivers Conserv 28:1431–1450. https://doi.org/10.1007/s10531-019-01732-9
Naylor GJP, Ryburn JA, Fedrigo O, Lopez A (2005) Phylogenetic relationships among the Major Lineages of Modern Elasmobranchs. In: Hamlett WC, Jamieson BGM (eds) Reproductive Biology and Phylogeny of Chondrichthyes: sharks, batoids, and chimaeras, vol 3. Science Publishers, Inc., Enfield, NH, pp 1–25
Naylor GJP, Caira JN, Jensen K, Rosana KAM, White WT, Last PR (2012) A DNA sequence-based approach to the identification of shark and ray species and its implications for global elasmobranch diversity and parasitology. Bull Am Mus Nat Hist 367:1–262. https://doi.org/10.1206/754.1
Pazmiño DA, van Herderden L, Simpfendorfer CA, Junge C, Donnellan SC, Hoyos-Padilla EM, Duffy CAJ, Huveneers C, Gillanders BM, Butcher PA, Maes GE (2019) Introgressive hybridisation between two widespread sharks in the East Pacific region. Mol Phylogenet Evol 136:119–127. https://doi.org/10.1016/j.ympev.2019.04.013
Petean FF, Naylor GJP, Lima SMQ (2020) Integrative taxonomy identifies a new stingray species of the genus Hypanus Rafinesque, 1818 (Dasyatidae, Myliobatiformes) from the Tropical Southwestern Atlantic. J Fish Biol 97:1120–1142. https://doi.org/10.1111/jfb.14483
Pincinato RBM, Gasalla MA, Garlock T, Anderson JL (2022) Market incentives for shark fisheries. Mar Policy 139:105031. https://doi.org/10.1016/j.marpol.2022.105031
Rigby CL, Dulvy NK, Barreto R, Carlson J, Fernando D, Fordham S, Francis MP, Herman K, Jabado RW, Liu KM, Marshall A, Pacoureau N, Romanov E, Sherley RB, Winker H (2019) Sphyrna lewini. The IUCN Red List of Threatened Species 2019: e.T39385A2918526
Robillard M, Séret B (2006) Cultural importance and decline of sawfish (Pristidae) populations in West Africa. Cybium 30:23–30
RStudio Team, RStudio (2022) RStudio: Integrated Development Environment for R. PBC, Boston, MA URL. http://www.rstudio.com
Sambrook J, Russell DW (2001) Molecular cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press, New York
Seidu I, van Beuningen D, Brobbey LK, Danquah E, Oppong SK, Séret B (2022a) Species composition, seasonality and biological characteristics of Western Ghana’s elasmobranch fishery. Reg Stud Mar Sci 52:102338. https://doi.org/10.1016/j.rsma.2022.102338
Seidu I, Brobbey LK, Danquah E, Oppong SK, van Beuningen D, Dulvy NK (2022b) Local ecological knowledge, catch characteristics, and evidence of Elasmobranch depletions in Western Ghana Artisanal fisheries. Hum Ecol 50(6):1007–1022. https://doi.org/10.1007/s10745-022-00371-z
Séret B, Naylor GJP (2016) Rhynchorhina mauritaniensis, a new genus and species of wedgefish from the eastern central Atlantic (Elasmobranchii: Batoidea: Rhinidae). Zootaxa 4138:291–308. https://doi.org/10.11646/zootaxa.4138.2.4
Simpfendorfer CA, Kyne PM (2009) Limited potential to recover from overfishing raises concerns for deep-sea sharks, rays and chimaeras. Environ Conserv 36:97–103. https://doi.org/10.1017/S0376892909990191
Simpfendorfer CA, Kyne PM, Noble TH, Goldsbury J, Basiita RK, Lindsay R, Shields A, Perry C, Jerry DR (2016) Environmental DNA detects critically endangered largetooth sawfish in the wild. Endanger Species Res 30:109–116. https://doi.org/10.3354/esr00731
Smart JJ, Chin A, Baje L, Green ME, Appleyard SA, Tobin AJ, Simpfendorfer CA, White WT (2016) Effects of including misidentified sharks in life history analyses: a case study on the Grey reef shark Carcharhinus amblyrhynchos from Papua New Guinea. PLoS ONE 11:e0153116. https://doi.org/10.1371/journal.pone.0153116
Spens J, Evans AR, Halfmaerten D, Knudsen SW, Sengupta ME, Mak SST, Sigsgaard EE, Hellström M (2017) Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: advantage of enclosed filter. Method Ecol Evol 8:635–645. https://doi.org/10.1111/2041-210X.12683
Thomsen PF, Kielgast J, Iversen LL, Møller PR, Rasmussen M, Willerslev E (2012) Detection of a Diverse Marine Fish Fauna using environmental DNA from seawater samples. PLoS ONE 7:e41732. https://doi.org/10.1371/journal.pone.0041732
Tillett BJ, Field IC, Bradshaw CJA, Johnson G, Buckworth RC, Meekan MG, Ovenden JR (2012) Accuracy of species identification by fisheries observers in a north Australian shark fishery. Fish Res 127–128:109–115. https://doi.org/10.1016/j.fishres.2012.04.007
Trégarot E, Catry T, Pottier A et al (2018) Évaluation des services écosystémiques du Banc d’Arguin, Mauritanie: rapport final. University of Portsmouth, CEE-M (Centre d’Economie de l’Environnement - Montpellier), IRD (Institut de Recherche pour le Développement), Nova Blue Environnement, IMROP (Institut Mauritanien de Recherches Océanographiques et des Pêches), FFEM (Fonds Français pour l’Environnement Mondial), PNBA (Parc national du Banc d’Arguin), AFD (Agence Française de Développement), BaCoMab. https://hal.archives-ouvertes.fr/hal-02091352
Trégarot E, Meissa B, Gascuel D, Sarr O, El Valy Y, Wagne OH, Kane EA, Bal AC, Haidallah MS, Fall AD, Dia AD, Failler P (2020) The role of marine protected areas in sustaining fisheries: the case of the National Park of Banc d’Arguin. Mauritania Aquac Fish 5:253–264. https://doi.org/10.1016/j.aaf.2020.08.004
Valadou B, Brêthes JC, Inejih CAO (2006) Observations biologiques sur cinq espèces d’Élasmobranches du Parc National Du Banc d’Arguin (Mauritanie). Cybium 30:313–322. https://doi.org/10.26028/cybium/2006-304-004
Valdés L, Déniz-González I (2015) Oceanographic and biological features in the Canary Current large marine ecosystem. IOC‐UNESCO, IOC Technical Series 115, Paris
Vianna GMS, Meekan MG, Ruppert JLW, Bornovski TH, Meeuwig JJ (2016) Indicators of fishing mortality on reef-shark populations in the world’s first shark sanctuary: the need for surveillance and enforcement. Coral Reefs 35:973–977. https://doi.org/10.1007/s00338-016-1437-9
Vilasboa A, Lamarca F, Solé-Cava AM, Vianna M (2022) Genetic evidence for cryptic species in the vulnerable spiny butterfly ray Gymnura altavela (Rajiformes: Gymnuridae). J Mar Biol Assoc United Kingd 102:345–349. https://doi.org/10.1017/S002531542200056X
Walker P, Cavanagh RD, Ducrocq M, Fowler SL (2005) Regional overviews. In: Fowler SL, Cavanagh RD, Camhi M, Burgess GH, Cailliet GM, Fordham SV, Simpfendorfer CA, Musick JA (eds) Sharks, rays and chimaeras: the Status of the Chondrichthyan fishes. Status survey, IUCN/ SSC shark specialist Group. IUCN, Gland, Switzerland and Cambridge, UK, pp 71–94
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species. Philos Trans R Soc Lond B Biol Sci 360:1847–1857. https://doi.org/10.1098/rstb.2005.1716
Westlund L, Charles A, Garcia SM, Sanders J (2017) Marine protected areas: Interactions with fishery livelihoods and food security. FAO Fisheries and Aquaculture Technical Paper No. 603, Rome, FAO
Wolff WJ, van der Land J, Nienhuis PH, de Wilde PAWJ (1993) Ecological studies in the Coastal Waters of Mauritania. Hydrobiologia 258:211–222
Yamamoto S, Masuda R, Sato Y, Sado T, Ara H (2017) Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci Rep 7:40368. https://doi.org/10.1038/srep40368
Yokota L, de Carvalho MR (2017) Taxonomic and morphological revision of butterfly rays of the Gymnura micrura (Bloch & Schneider 1801) species complex, with the description of two new species (Myliobatiformes: Gymnuridae). Zootaxa 4332:1–74. https://doi.org/10.11646/zootaxa.4332.1.1
Zhang S, Zhao J, Yao M (2020) A comprehensive and comparative evaluation of primers for metabarcoding eDNA from fish. Method Ecol Evol 11:1609–1625. https://doi.org/10.1111/2041-210X.13485
Acknowledgements
Research permits (V/L nº160/PM/MSGG) were granted by the Ministry of Environment and Sustainable Development (Ministère de l’Environnement et du Développement Durable). The Pew Charitable Trusts supported this workthrough a Pew Fellowship in Marine Conservation awarded to EAS and RWJ. We additionally thank Projects MARAFRICA-AGA-KHAN/540316524/2019 (Aga Khan Foundation and Fundação para a Ciência e a Tecnologia, including UIDB/P/04326/2020), Projet Suivi des Tortues Marines (Partenariat Régional pour la Conservation de la zone côtière et Marine en Afrique de l’Ouest - PRCM), the Shark Conservation Fund (a project of Rockefeller Philanthropy Advisors), TROPIBIO (EU-H20202 854248), and LinnéSys: Systematics Research Fund of the Linnean Society of London and the Systematics Association for funding various aspects of this research. We also thank the Parc National du Banc d’Arguin (PNBA) and the Institut Mauritanien de Recherches Océanographiques et de Pêches (IMROP) for supporting this project. We are especially grateful to Mamadou Abdoul Ba, Mohammed Bourweiss, and Alioune Niang for their in-country assistance and support, as well as the various PNBA and IMROP members of staff who have facilitated this research, including Sall Amadou, Oumar Samba Wone, Abdoulaye Soumare, Ahmed Boubout and Ahmed Etfagha. For support during field sampling we thank PNBA captain Mohamed Salem and Imraguen guide Mohamed Chedad, the Imraguen fishers and community including captain Mohamed Ayoub, IMROP adviser director Khallahi Brahim and the crew of R/V Amrigue for support during the 2020 oceanographic sampling. We are also thankful to Stefano Mariani, Gareth A. Pearson, Gavin Naylor, Joyce Velos, and Marta Valente for providing help and critical feedback during various stages of this project.
Funding
Open access funding provided by FCT|FCCN (b-on). This work was supported by The Pew Charitable Trusts through a Pew Fellowship in Marine Conservation awarded to Ester A. Serrão and Rima W. Jabado. Partial financial support was received from Projects MARAFRICA-AGA-KHAN/540316524/2019 (Aga Khan Foundation and Fundação para a Ciência e a Tecnologia, including UIDB/P/04326/2020), Projet Suivi des Tortues Marines (Partenariat Régional pour la Conservation de la zone côtière et Marine en Afrique de l’Ouest - PRCM), the Shark Conservation Fund (a project of Rockefeller Philanthropy Advisors), TROPIBIO (EU-H20202 854248), and LinnéSys: Systematics Research Fund of the Linnean Society of London and the Systematics Association.
Author information
Authors and Affiliations
Contributions
EAS, RWJ, AV, and CHS contributed to the study conception and design. Material preparation and data collection were performed by CHS, EAS, RWJ, ES, and CYG. CHS, AV, and LC performed laboratory work and analysis. CHS wrote the main manuscript text and prepared all figures. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose and have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de la Hoz Schilling, C., Jabado, R.W., Veríssimo, A. et al. eDNA metabarcoding reveals a rich but threatened and declining elasmobranch community in West Africa’s largest marine protected area, the Banc d’Arguin. Conserv Genet 25, 805–821 (2024). https://doi.org/10.1007/s10592-024-01604-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-024-01604-y