Abstract
Despite their high abundance and species richness, tilapiines have been compromised by various factors especially overfishing, climate change, and uncontrolled fish transfers and introductions. Fish introductions have negatively impacted native tilapiine populations through competition, predation, hybridization, and introgression compromising their genetic integrity. The hybridization levels of different tilapiines in the Lake Victoria basin remain relatively understudied. The study utilized nuclear microsatellite and mitochondrial DNA (mtDNA) genetic markers to investigate hybridization signals and compare the genetic diversity of different tilapiines in Lake Victoria, Kenya, using next-generation sequencing. Low levels of hybridization from Oreochromis niloticus into other Oreochromis species were detected by Bayesian clustering analysis and principal coordinate analysis (PCoA). The results contribute to the need for conservation measures for these fish species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The African cichlids, particularly tilapiines have been widely distributed in the temperate, tropics, and sub-tropic natural water bodies including the great lakes of Africa. Most of the African lakes especially Tanganyika, Malawi, and Victoria are depicted as centers of adaptive radiation for the cichlids (Salzburger and Meyer 2004; Meyer et al. 2015). Cichlids are considered important model species for evolutionary biological research due to their morphological diversity as well as species richness with more than 1600 species and at least 20 genera (Salzburger and Meyer 2004). Among the cichlids, the tilapiines which comprise different genera of the species Oreochromis niloticus, Oreochromis mossambicus, and Oreochromis aureus have gained significant scientific attention with most studies focussing on strain selection, genetic improvements, and aquaculture (Eknath and Hulata 2009; Firmat et al. 2013; Kwikiriza et al. 2023). Aquaculture production in Africa dates to the 1940 and 1950s with Egypt leading followed by Nigeria, Uganda, Ghana, and Kenya in that order (FAO 2022). Tilapia production in Kenya contributes at least 60.2% of the total fish production with 50.99% from aquaculture systems like ponds and cages while 9.21% is derived from natural water bodies mostly rivers and lakes (Opiyo et al. 2018). Despite the conservation measures developed for sustainable utilization of capture fisheries, the sector has kept at crossroads due to higher fishing pressure that comes along with catching immature fishes, climate change effects and un-controlled fish transfers, etc. (Eknath and Hulat 2009; Njiru et al. 2010).
The introduction of tilapiines in East African freshwater bodies dates back to the 1950s, with Kenya being a hotspot (Mwanja et al. 2010, Okwiri et al. 2019). Many intentional and unintentional cases of species introductions and transfers have been reported in Kenya (Njiru et al. 2010; Ndiwa et al. 2014). Kenya’s freshwater bodies have so far had at least 14 fish introductions described and documented (Hickley et al. 2002). Among these introductions, six fish intentional introductions have taken place in Lake Victoria, seven in Lake Naivasha and one has occurred in Lake Baringo (Ogutu-Ohwayo and Hecky 1991; Hickley et al. 2002). All these introductions have comprised Lates niloticus, Cyprinus carpio, Coptodon zillii, Oreochromis niloticus, Oreochromis leucostictus, and Oreochromis melanopleura (Ogutu-Ohwayo and Hecky 1991; Ndiwa et al. 2014).
Unintentional/accidental cases of species transfers by fish farmers have also been reported in Kenya (Ndiwa et al. 2014). For example, due to poor farm management and designs, most of these earthen pond farms are located near the water bodies, and in cases of heavy rains, the farms flood, and fish end up escaping into the natural water bodies thus mixing with the wild species affecting the genetic pool of the wild stock (Munguti et al. 2014; Ndiwa et al. 2014). Studies show that in 2013, several Oreochromis niloticus stocks found their way into Lake Victoria from nearby fish farms due to heavy floods (Ndiwa et al. 2014; Deines et al. 2014).
The introduction of fish species in the Lake Victoria basin, like elsewhere accounts for positive and negative implications. For example, the introduction of Coptodon zillii was purposely to fill the niche of macrophytophage while Oreochromis niloticus and Oreochromis leucostictus were introduced to boost fisheries of the native tilapiines that had reduced because of overfishing (Ndiwa et al. 2014). Introductions of Lates niloticus were purposely to boost the fishery as it grows into bigger sizes after predating on the bony and small, but abundant, haplochromines (Mwanja et al. 2010).
On the other hand, fish introductions have led to the disappearance of the native species in the natural water bodies due to competition for resources and hybridization levels (Hickley et al. 2002; Angienda et al. 2011; Shechonge et al. 2018; Blackwell et al. 2020; Kariuki et al. 2021). Oreochromis niloticus, an invasive species has displaced many native species and could have altered their ecology (Champneys et al. 2021). Many authors have stated that the invasion of Oreochromis niloticus in locations naturally inhabited by congeneric species often leads to hybridization (Tibihika et al. 2020; Kariuki et al. 2021; Champneys et al. 2021). For example, the introduction of Oreochromis niloticus in Lake Victoria is likely to have contributed to the decline and expatriation of the native species mainly Oreochromis variabilis and Oreochromis esculentus from the main lake, and these species are currently found in some satellite lakes of Kanyaboli, Bisina and Mburo (Mwanja et al. 2010; Shechonge et al. 2018; Tibihika et al. 2020; Blackwell et al. 2020; Kariuki et al. 2021). Similarly, Mwanja et al. (2010) observed that Oreochromis variabilis was not found in Lake Sare and attributed this to overfishing. In Lake Victoria, studies by Mwanja et al. (2010) observed hybrids resulting from Oreochromis niloticus with Oreochromis esculentus in the satellite lakes concluding that no pure strains of Oreochromis esculentus existed.
Shechonge et al. (2018) report cases of hybridization between the introduced Oreochromis niloticus and the native tilapiines in the Mindu reservoir of Ngerengere River which is part of the Ruvu system in the Eastern part of Tanzania. Angienda et al. (2011) discovered minimal nuclear gene transfer from Oreochromis niloticus to Oreochromis esculentus in Lake Kanyaboli and Namboyo, Kenya. This was attributed to the introduction of Oreochromis niloticus in Lake Kanyaboli with its high hybridization levels. The study by Deines et al. (2014) found that the native cichlids Oreochromis macrochir and Oreochromis andersonii hybridize in the presence of Oreochromis niloticus. The two native species do not naturally hybridize when in sympatry, suggesting that the presence of the non-native species facilitates hybridization events (Deines et al. 2014).
Despite these emerging concerns arising from the hybridization of Oreochromis niloticus with congeneric species, the extent of this in its native range has received less attention in Lake Victoria. Moreover, incidences of hybridization complicate the distinction between the Oreochromis species, and this might affect management and conservation measures aimed at ensuring the sustainability of the resources (Tibihika et al. 2020). This study employed two markers: microsatellites and mitochondrial DNA (mtDNA) to provide a comprehensive understanding of the genetic structure of these tilapiines than using one molecular marker. This is because microsatellites are highly polymorphic and are co-dominantly inherited (Labiros et al. 2022), whereas mtDNA has uniparental inheritance (maternal) and does not have recombination (Parakatselaki and Ladoukakis 2021).
Although there is prevailing research on the genetic structure, diversity, differentiation, etc. in East Africa, many of these studies used either microsatellite markers or mtDNA markers to study tilapiines in the region. However, the microsatellite genetic markers used are known to reveal significant weaknesses/limitations, such as size homoplasy (Mwanja et al. 2010; Tibihika et al. 2020).
Therefore, the present study used microsatellite markers and mtDNA using next-generation sequencing to elucidate the degree of admixtures and hybridization levels of different Tilapiines collected from Lakes; Victoria and Sare. Next-generation sequencing (NGS) has proven to be the best approach as it minimizes size homoplasy, which is one of the constraints of the traditional simple sequence repeats (SSR) fragment length approach (Tibihika et al. 2020). The results contribute to the knowledge of the genetic variation found within both Oreochromis species in their native range and help in defining future stocks for aquaculture.
Materials and methods
Sample collection
A total of 322 fish samples were captured using experimental gill nets with the help of fishermen from Lake Victoria and its satellite lakes. Each sample had at least 150 g for easy differentiation and classification. The sampling sites were; Mbita, Dunga, Luanda Nyamasare, Usenge, Usoma, Siungu, Seka Bay and Lake Sare (Fig. 1). All these sites are in the Lake Victoria basin, Kenya (Fig. 1). The targeted tilapiine species in all sites included; Oreochromis esculentus, Oreochromis leucostictus, Oreochromis niloticus, and Coptodon zillii (Table 1). Samples of; Oreochromis leucostictus, Oreochromis niloticus, and Coptodon zillii were obtained from Lake Victoria while Oreochromis esculentus and Oreochromis leucostictus were collected from lake Sare. Animal rights were observed during the collection of fish samples. The collected fish muscle or fin tissue was preserved in 98% ethanol for subsequent genotyping. Before airlifting the samples to Austria, official authorization was granted from the respective bodies responsible for fisheries corresponding to each region.
Microsatellite and mtDNA genotyping
DNA extraction followed the protocol of Tibihika et al. (2019) using magnetic beads (MagSi-DNA beads-MagnaMedics) and a magnetic separator SL-MagSep96 (Steinbrenner, Germany) with an improved MagSi-DNA Vegetal kit protocol. DNA quality was verified using 1.5% agarose gel and later visualized using a trans-illuminator system prior to amplification. Polymerase Chain Reaction (PCR) amplification was carried out using 43 previously developed microsatellites (SSR) for Oreochromis niloticus populations from East Africa (Tibihika et al. 2019). The selected primers were then divided into four multiplexes and used to prepare SSR genotyping by amplicon sequencing libraries (SSR-GAS) based on the procedures of Kariuki et al. (2021). PCR reactions were carried out in a 5.0 µl total volume containing 1.0 µl of the genomic DNA, 1.0 µl of primer mix, 2.5 µl of Master mix (Qiagen Multiplex PCR Kit; Qiagen; Netherlands), and 0.5 µl of autoclaved water. The conditions were as follows: initialization at 95 °C for 15 minutes, followed by denaturation for 30 cycles at 95 °C for 30 seconds, annealing at 55 °C for one minute, and elongation for one minute at 72 °C and the last extension steps at 72 °C for 10 minutes. The PCR products were pooled sample-wise, cleaned using AMPure magnetic beads, and subsequent indexing was performed following Tibihika et al. (2019). The Individually indexed samples were pooled and used for a paired-end 300 bp sequencing run on an Illumina MiSeq at the Genomics Service Unit at Ludwig Maximilian Universität, München, Germany.
For mtDNA genotyping, primers specific for Tilapia species were constructed by comparing existing mitochondrial genomes. Three primers were used to amplify parts of the COI region. Amplification and sequencing were done following the same protocols used in SSR genotyping.
Sequence analysis, genotyping, and allele calling of microsatellite and mtDNA
Products from Illumina sequencing were subjected to quality check using FastQC version 0.11.9 and trimmed using Trimmomatic version 0.39 (Bolger et al. 2014) to remove adapters and poor-quality regions. Further steps including merging, demultiplexing, and allele calling were done using the Python scripts described in Curto et al. (2019). Given the haploid nature of mtDNA, if more than one allele was found per individual, the one with the higher read count was kept. The sequences were then aligned with MAFFT (Katoh and Standley 2013) using the global alignment option and primers were removed manually.
Statistical analysis
Microsatellite data
The SSR codominant matrix was analyzed with GenAlEx v. 6.503 (Peakall and Smouse 2006) to assess several variability measures per population, including the average number of alleles per locus (Na), the effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), F-statistics (FST) and Shannon’s information index (I). The same program was used to estimate paired-wise FST values across all populations and also produce absolute genetic distances among the tilapiines that were visualized in a principal coordinate analysis (PCoA). Genetic structure and differentiation between populations were further inspected in two ways. First, by visualizing Nei’s genetic distances (Schneider et al. 2000) with an unweighted pair group method with arithmetic mean (UPGMA) dendrogram as executed in Populations v.1.2.32 (Langella 1999). The support values were estimated by performing 1000 bootstrap replicates with loci resampling.
Second, by using STRUCTURE v. 2.3.4 (Hubisz et al. 2009) program to analyse the genetic clustering of samples. It involved 13 iterations with K set from 1 to 10 and 100,000 generations after a burn-in period of 10, 000. This was done for datasets consisting of all tilapiines and their respective populations.
MtDNA data
The 275 sequences obtained during this study were aligned and compared with published GenBank sequences that showed a high similarity to our sequences during BLASTN searches. All sequences were analyzed for four recognized tilapiine species which included (Oreochromis niloticus GenBank: GU477625, GU477628, MG728092, MN384747, MG728093, MG728006, and MG728058), (Coptodon zillii Genbank: AF296503, AF296504, EU163713, EU163722, KJ925088, KY465484, and MH64435), Oreochromis esculentus Genbank: AF296480, KM654981, MG922062 and MH717437 and Oreochromis leucostictus Genbank: EF016702, KX757687, KX757703, KX767805, KX767823, and MG922061). mtDNA control region sequences were aligned with the software BioEdit v.7.0 (Hall 1999) and checked manually. To determine levels of extant genetic diversity, the number of polymorphic sites and mtDNA haplotypes, haplotype diversity (Hd), Tajima’s D, and nucleotide diversity (π) were calculated using DnaSP Version: 6.12 (Librado and Rozas 2009) for each species as well as the entire pooled population of each species. PopArt was used to construct the haplotype network to investigate the phylogenetic relationships among the mtDNA haplotypes of each species (Leigh and Bryant 2015). Ambiguous connections in the haplotype network generated by PopArt were resolved following the criteria outlined in Crandall and Templeton (1993).
Results
Genetic diversity of tilapiines using microsatellite markers
Based on heterozygosity and number of alleles (Na), the present results show that the populations of Oreochromis niloticus were more genetically diverse than the other tilapiines (Table 2a). Furthermore, the heterozygosity of Oreochromis niloticus populations from different beaches was evaluated and the results show that the expected heterozygosity is higher than the observed heterozygosity (Table 2b); the samples from Dunga beach showed higher heterozygosity than other beaches (Table 2b).
Population differentiation using pairwise FST values
Pairwise FST values were equal with Oreochromis niloticus vs. Oreochromis leucostictus having the highest pairwise comparison (0.36) followed by Oreochromis niloticus and Oreochromis esculentus (0.35) while Oreochromis leucostictus and Coptodon zillii had the lowest FST values (0.067) within populations (Table 3).
Genetic structure and signs of admixture of theOreochromisspecies.
The PCoA for the Oreochromis species formed two main clusters with populations of Oreochromis niloticus forming an independent cluster from the remaining species (Fig. 2a). In this case, at least five samples of Oreochromis niloticus were found in the cluster containing Oreochromis leucostictus and Oreochromis esculentus (Fig. 2a). Two main subclusters are also observed in the cluster containing Oreochromis leucostictus and Oreochromis esculentus populations. One subcluster contains more samples of Oreochromis leucostictus with a few Oreochromis esculentus while the second subcluster is mixed with both Oreochromis esculentus, Oreochromis leucostictus and Oreochromis niloticus populations (Fig. 2a).
Populations of Oreochromis niloticus and Oreochromis leucostictus formed two main clusters; cluster one containing Oreochromis niloticus and another one containing Oreochromis leucostictus. Few samples of Oreochromis niloticus were observed in a cluster containing Oreochromis leucostictus (Fig. 2b). A sub-cluster was observed containing Oreochromis niloticus populations (Fig. 2b).
The PCoA results of the Oreochromis species from Lake Sare formed intermediate clusters indicating admixtures of Oreochromis esculentus and Oreochromis leucostictus (Fig. 2c). Sample populations of Oreochromis leucostictus formed a sub-cluster (Fig. 2c).
Oreochromis leucostictus samples from Lakes; Victoria and Sare formed two clusters that are close to each other. Samples from Lake Sare formed a sub-cluster that was far from the two main clusters (Fig. 2d). The PCoA was conducted to differentiate the populations of Oreochromis niloticus from different beaches using the first three axes.
The PCoA showed two clear groups: Oreochromis niloticus populations from Usenge, Mbita, Siungu, Luanda and Seka-Bay formed one cluster while populations from Dunga and Usoma formed another cluster (Fig. 2e). In cluster one, populations from Usenge and Siungu were very close compared to populations from Mbita and Luanda (Fig. 2e).
The UPGMA dendrogram is generally consistent with the PCoA in Fig. 2a and demonstrates similar patterns (Fig. 3). Among the tilapiines, the largest separation was between Oreochromis niloticus populations and other tilapiines (Oreochromis esculentus, Coptodon zillii, and Oreochromis leucostictus). Consistently, Oreochromis niloticus populations clustered together (bootstrap > 80). Oreochromis leucostictus and Oreochromis esculentus were the most genetically similar populations (Fig. 3).
Considering Oreochromis niloticus populations, the UPGMA dendrogram is generally congruent with the PCoA in Fig. 2e (Fig. 3). Consistently, samples from Siungu beach showed the most divergence from other beaches followed by Dunga beach. Samples from Luanda Nyamasare and Usenge showed similar distinctions as are Mbiita and Usoma. Samples from Seka Bay are more like Usenge and Luanda Nyamasare (Fig. 3).
The STRUCTURE v. 2.3.4 outputs observed two main clusters among the Oreochromis species which are congruent with PCoA and UPGMA analyses in figures: 2a and 3 respectively (Fig. 4a). Similarly, the Oreochromis niloticus and Oreochromis leucostictus formed two independent clusters (Fig. 4b) and this aligns with the PCoA results in Fig. 2b.
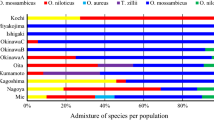
The populations of Oreochromis niloticus from Dunga and Luanda, Seka-Bay and Usenge showed some degree of admixture while samples from Mbita and Usoma beaches showed a pure population (Fig. 4c). Generally, all the structure outputs were congruent with the PCoA outputs from GenAlex.
Genetic diversity and population differentiation using mtDNA markers
After the alignment of all 299 sequences (275 newly obtained and 24 downloaded from GenBank), 23 haplotypes were identified (Table 4; Fig. 5). The dataset contained 84 polymorphic sites and 82 parsimony informative sites. The haplotype diversity (Hd) for the four species ranged from 0.00 to 1.00 (Table 4) with Oreochromis niloticus having the highest Hd value (Hd: 0.76) followed by Coptodon zillii (Hd: 0.30) while the lowest (Hd: 0.00) was found in Oreochromis esculentus where only one haplotype was produced, indicating the presence of allelic indifference. Additionally, nucleotide diversity (π) for the four species ranged from 0.00 to 0.04, with Oreochromis niloticus and Coptodon zillii having a similar value (π = 0.04) while Oreochromis esculentus had the lowest value (π = 0.00) (Table 4).
Population differentiation of the tilapiines
When the collected samples were compared with sequences from the gene bank, 23 haplotypes were observed with Oreochromis niloticus having seven monomorphic haplotypes (4,5,9,10,11,12 and 13) (Fig. 5). In addition, Hap4 and Hap2 had greater dominance in Oreochromis niloticus compared to other haplotypes. Five (9,10,11,12 and 13) of the seven haplotypes for Oreochromis niloticus were private or singleton haplotypes which generally indicated the different levels of genetic variability in the entire data set. The most widespread haplotypes were Hap1 and Hap2, with Hap1 being the most widely distributed across the entire data set (5 species; Oreochromis niloticus, Oreochromis leucostictus, Oreochromis esculentus and Coptodon zillii collected from the field and Oreochromis leucostictus from the GenBank). Additionally, Hap2 was shared by Oreochromis niloticus collected from the field and Oreochromis niloticus from the GenBank with field samples showing higher dominance. Oreochromis niloticus, Oreochromis leucostictus, and Oreochromis esculentus shared haplotype 3. Haplotypes 15 and 16 were shared by Oreochromis niloticus and Oreochromis leucostictus samples (Fig. 5).
Discussion
Despite the economical and conservational importance of fisheries resources, they have been severely compromised by human-induced activities that affect the sustainable utilization of such resources (Eknath and Hulata 2009; Kwikiriza et al. 2023). These anthropogenic activities include; habitat destruction, overfishing, and unregulated fish transfers (Eknath and Hulata 2009). They have altered the natural genetic structure of different fish species especially Oreochromis species through admixture and hybridization (Tibihika et al. 2020). Therefore, it’s essential to understand the extent of genetic divergence of these fishes as this helps in the efficient management of wild fish populations and for aquaculture activities. This information can be achieved through the differentiation of the tilapiines using highly informative genetic markers especially microsatellite and mtDNA genotyping.
Genetic diversity and differentiation of Oreochromis niloticus populations
The populations of Oreochromis niloticus were highly genetically diverse both at mtDNA and microsatellite loci compared to other species. Heterozygosity compares the amount of genetic variation within different populations (Gu et al. 2014; Kajungiro et al. 2019). In the current study, the overall observed heterozygosity was lower than the expected heterozygosity for Oreochromis niloticus populations. Different studies have shown lower observed heterozygosity and attributed this to factors like the presence of null alleles, sample size, and inbreeding levels of the different Oreochromis species as well as Wahlund effect (D’amato et al. 2007; Garnier-Géré and Chikhi 2013; Gu et al. 2014; Kajungiro et al. 2019). For example, studies by Kajungiro et al. (2019) observed lower heterozygosity in Oreochromis niloticus populations and attributed this to the small sample size which was used to infer the findings. Similarly, Gu et al. (2014) found that observed heterozygosity in six populations in the primary rivers of Guangdong province was lower than the expected heterozygosity and attributed it to inbreeding at many loci and in all of the Oreochromis niloticus populations. Therefore, the current lower heterozygosity among the Oreochromis niloticus could be partially attributed to the occurrence of non-random mating among the populations since the sample size of the population in each beach was at least 30. This indicates that the Oreochromis niloticus population is not affected by smaller amounts of genetic drift as populations are generally larger thus having higher effective population size and consequently higher genetic diversity (Martinez et al. 2018). Additionally, the low heterozygosity levels could be due to the Wahlund effect (Garnier-Géré and Chikhi 2013) which indicates that observed heterozygosity is reduced as populations diverge in an aquatic environment.
In comparison with other species, Oreochromis niloticus showed a higher heterozygosity which generally translates to higher genetic diversity than other species. The study utilized 136 samples of Oreochromis niloticus collected from different beaches which were far larger than other species: 60, 57, and 30 for Oreochromis leucostictus, Coptodon zillii, and Oreochromis esculentus respectively. This indicates that the Oreochromis niloticus population is not affected by smaller amounts of genetic drift as populations were generally bigger thus having higher effective population size and consequently higher genetic diversity (Martinez et al. 2018).
The admixtures observed among the Oreochromis niloticus populations as evidenced by PCoA and Structure outputs is attributed to the chances/instances of uncontrolled movement/transfer of fish from one location to another as well as increased escapees from aquaculture (Ndiwa et al. 2014) since most of the sampled beaches are close to each other and some directly connected to the streams and rivers. To further explore the dynamics of admixture within these populations, the distribution of Oreochromis niloticus haplotypes in the study was considered. All the haplotypes of Oreochromis niloticus are close to each other with less than five mutational changes indicating the higher levels of admixtures. All these confirm that there is human-mediated gene flow between Oreochromis niloticus populations in the sampled beaches.
It is also expected that aquaculture activities might be contributing to the observed gene flow between the populations on these beaches. The increased aquaculture activities in the Rift Valley region have enhanced fish transfer from one drainage system to another allowing mixing between populations and or species (Ndiwa et al. 2014). The Economic stimulus program (ESP) introduced in 2009 by the Kenyan government has tremendously led to the establishment of many fishponds; some are constructed near wetlands, streams, rivers, and lakes (Munguti et al. 2014; Opiyo et al. 2018). During the heavy rains, the ponds get flooded leading to fish escaping into Lake Victoria. Most of these ponds are not isolated from streams and wetlands, thus farmed fish can easily escape and hybridize with autochthonous (Angienda et al. 2011; Ndiwa et al. 2014).
The current mtDNA results show a relatively higher haplotype diversity in Oreochromis niloticus compared to other tilapiines as more than five monomorphic haplotypes were observed. The current haplotype diversity for Oreochromis niloticus aligns with the results (Hd: 0.800 in Oreochromis niloticus) obtained by Abdel-Hamid et al. (2014). Since the introduction of Oreochromis niloticus dates back to the 1950s in Lake Victoria, such time is enough for an introduced population to establish genetic divergence due to ecological tolerance and biological fecundity (Aloo 2003; Angienda et al. 2011; Firmat et al. 2013; Tibihika et al. 2020). Tibihika et al. (2020) observed a higher diversity of Oreochromis niloticus and attributed this to the admixtures originating from several lineages as a result of multiple fish stockings. Therefore, the higher haplotype diversity of Oreochromis niloticus populations could be attributed to the widespread introductions and distributions of Oreochromis niloticus to different geographical areas within East African water bodies, especially Lake Victoria. Additionally, the strong founder effects during colonization into new habitats may also contribute to the substantial genetic differentiation among the populations of Oreochromis niloticus.
The low genetic diversity of the other tilapiines in the study could be due to the hybridization levels with Oreochromis niloticus (Ndiwa et al. 2014). Haplotype 2 (Hap2) which was shared by Oreochromis niloticus from the field and Oreochromis niloticus sequences from the GenBank suggested that all these populations might be originating from similar maternal ancestors (Jiang et al. 2019). Therefore, the higher haplotype diversity indicates that the populations contain an abundant genetic resource for subsequent use in breeding or conservational measures.
Various approaches using both multivariate analysis; Principal Coordinates Analysis (PCoA) and Bayesian clustering algorithms (STRUCTURE) were used in this study to evaluate the genetic structure of the Oreochromis species. The results of PCoA showed two main clusters with Oreochromis niloticus populations forming an independent cluster while other species (Oreochromis esculentus and Oreochromis leucostictus) formed another cluster. Such independent clusters could be explained by the differences in mutation and selection associated with the evolutionary history of populations as well as drift and migration linked with the effects of fragmentation of populations and their demographic background (Martinez et al. 2018). For example, Oreochromis niloticus is primarily a phytoplankton feeder and dominates areas of dense algal stocks (Ndiwa et al. 2014; Laurent et al. 2020; Kwikiriza et al. 2023) while Oreochromis esculentus and Oreochromis leucostictus prefer habitats near papyrus fringes in littoral, shallow muddy bays, and lake inlets (Laurent et al. 2020). Probably such differences in habitat isolation contribute to the differences in PCoA and Structure outputs. Similarly, eight individuals of Oreochromis niloticus populations appeared in the cluster containing Oreochromis esculentus and Oreochromis leucostictus indicating that these individuals had some degree of admixtures. The presence of Oreochromis niloticus populations in the other cluster could be due to the potential misclassification of the species since admixed individuals resemble more Oreochromis niloticus and therefore they may have been misclassified (Angienda et al. 2011; Kariuki et al. 2021).
Oreochromis species from Lake Sare
Numerous studies on satellite lakes of the Victoria basin have led to the discovery of new fish species richness and genetic diversity which have not yet been sampled in the main Lake Victoria (Abila et al. 2008; Angienda et al. 2011). Sare; a satellite lake provides refugia for different fish species. The lake is connected to main Lake Victoria by extensive Yala papyrus swamps that are anoxic to invasive predators like Nile perch and prevent entry of the Oreochromis niloticus species (Abila et al. 2008). In the current study, the obtained low heterozygosity of Oreochromis leucostictus and Oreochromis esculentus suggests low genetic variability for these species (Kajungiro et al. 2019: Kariuki et al. 2021) and could be attributed to bottlenecks caused by fast reduction of population size due to overfishing. Similarly, the haplotype and nucleotide diversity of the Oreochromis leucostictus and Oreochromis esculentus were lower (Hd = 0.03; π = 0.00 and Hd = 0.00; π = 0.00) respectively. Lake Victoria has been experiencing high fishing pressure and species introductions leading to changes in the structure of the freshwater habitats, biodiversity, genetic structure, composition, and the productivity of the associated biota (Matsuishi et al. 2006). All these have led to the loss of genetic diversity and the disappearance of native fish stocks like Oreochromis esculentus from the main Lake and such species can only be found in the satellite lakes like Sare and Kanyaboli (Mwanja et al. 2010; Angienda et al. 2011; Kariuki et al. 2021). Contrary to our findings, studies by Angienda et al. (2011) obtained a higher observed heterozygosity in Lake Kanyaboli compared to present findings where the observed heterozygosity was lower in Lake Sare. The differences could be attributed to the types of microsatellite markers used, maybe the markers used by Angienda et al. (2011) were more variable than the markers used in the current study. Secondly, Lake Sare being smaller than Kanyaboli limits the population size of the fish fauna thus low genetic diversity (Aloo 2003).
In the present study, the PCoA results showed intermediate clustering of the two Oreochromis species. The intermediate clustering could be associated with hybridization which makes the species close to each other as they occupy similar ecosystems (Angienda et al. 2011; Laurent et al. 2020; Kariuku et al. 2021). Studies show that Oreochromis leucostictus and Oreochromis esculentus prefer habitats near papyrus fringes in littoral, shallow muddy bays, and lake inlets (Laurent et al. 2020). Therefore, this enhances their ability to randomly mate thus admixtures among the species. This pattern of phylogenetically intermediates ‘mixed’ populations observed in the study provides strong evidence for the occurrence of hybridization and indicates the direction of introgression.
Oreochromis leucostictus
In the present study, the distinct populations of Oreochromis leucostictus from Sare and Victoria could be attributed to the physical isolation created by the main road and Yala swamp. The two lakes are separated by a natural wetland, and the eco-physiological properties of these two lakes are different, which could also limit gene flow between the two populations by local adaptation or physiological barriers (Crispo and Chapman 2008). Although Oreochromis leucostictus from Lake Victoria and Sare is generally differentiated based on the PCoA output, some samples occupied an intermediate position. Since these lakes are so close to each other and only separated by the main road and Yala swamp (Aloo 2003), it is clearly possible that multiple stockings/ fish transfers might be happening or happened which contributes to the observed gene pool indicated by the intermediate populations. Oreochromis leucostictus populations of Lake Sare formed a subcluster and this is an indication that a pure stock still exists in the lake.
Hybridization between different species as a result of introductions
While hybridization events often occur among tilapiines following non-native species introductions into the natural environment, cases of hybridization between sympatric indigenous species are limited (Shechonge et al. 2018). In the present study, hybridization levels can be evidenced by the presence of the individuals of Oreochromis niloticus in a cluster containing Oreochromis leucostictus and the occurrence of haplotypes shared by Oreochromis niloticus and Oreochromis lecucostictus as indicated in other studies (Mwanja et al. 2010; Angienda et al. 2011; Deines et al.2014; Shechonge et al. 2018).
The current mtDNA results reveals that haplotype; 3, 15, 16, and 17 are shared among Oreochromis niloticus and Oreochromis leucostictus suggesting mtDNA introgression (Ndiwa et al. 2014) with a possibility of genetic admixture at the population phase. Other studies have indicated that Oreochromis niloticus has hybridized with other Oreochromis species (Deines et al. 2014; Blackwell et al. 2020; Diedericks et al. 2021). Elsewhere, studies by Ndiwa et al. (2014) observed low hybridization levels between Oreochromis niloticus and Oreochromis leucostictus in Lake Naivasha Kenya. He also indicated that the haplotype of Oreochromis leucostictus from Lake Naivasha had been previously described to occur in Oreochromis niloticus from the Lake Baringo population. Recent studies in the Lake Edward-George system indicate that hybridization between Oreochromis niloticus and Oreochromis leucostictus is happening though rarely (Diedericks et al. 2021). A study by Deines et al. (2014) found that the native cichlids Oreochromis macrochir and Oreochromis andersonii hybridize in the presence of Oreochromis niloticus. The two native species do not naturally hybridize when in sympatry, suggesting that the presence of the non-native species facilitates hybridization events (Deines et al. 2014). In Tanzania, studies by Blackwell et al. (2020) observed introgression levels between Oreochromis niloticus and Oreochromis korogwe as well as hybrids between Oreochrmois urolepis and Oreochromis korogwe. They attributed this to the introduced Oreochromis niloticus populations in Lake Nambawala that hybridized with the native species (Shechonge et al. 2018; Blackwell et al. 2020). Similarly, the genetic differentiation of the tilapiines based on the FST values indicates that Oreochromis niloticus is closer to Oreochromis leucostictus. This perhaps implies that gene flow is prominent among populations of Oreochromis niloticus with Oreochromis leucostictus and Oreochromis esculentus without physical barriers because of unregulated fish transfers and aquaculture activities (Shechonge et al. 2018; Laurent et al. 2020). Therefore, the present results contribute further evidence to the hypothesis that there is a low level of hybridization from Oreochromis niloticus into other Oreochromis species which likely threatens the conservation of these species.
Conclusion
The presence of Oreochromis niloticus populations in other clusters indicates some degree of admixture. The results confirm the hypothesis that there is a low level of introgression from Oreochromis niloticus.
The genetic diversity and structure of Oreochromis niloticus populations from different beaches can be explained by their life history and geographical distribution. The close clustering of Usenge and Siungu, Mbita and Luanda populations, and distinct separation of Dunga, suggests that these could be pure populations without admixture.
The current findings point out the need for a comparative study on the main lakes in East African countries to understand the extent of hybridization of tilapiines in these natural environments. This would provide a broader knowledge of genetic diversity that could be used when designing conservation measures.
Data Availability
All data analyzed for this study are part of this article.
References
Abdel-Hamid ZG, Heba AM, El-Kader A, Aboelhassan DM, Mahrous KF (2014) Genetic diversity in egyptian tilapia species using PCR-RFLP of D-loop mitochondrial DNA gene. Res J Pharm Biol ChemSci 5(6):469–475
Abila R, Salzburger W, Ndonga MF, Owiti DO, Barluenga M, Meyer A (2008) The role of the Yala swamp lakes in the conservation of Lake Victoria region haplochromine cichlids: evidence from genetic and trophic ecology studies. Lake Reserv Manag 13(2):95–104. https://doi.org/10.1111/j.1440-1770.2008.00366.x
Aloo PA (2003) Biological diversity of the Yala Swamp lakes, with special emphasis on fish species composition, in relation to changes in the Lake Victoria Basin (Kenya): threats and conservation measures. Biodivers Conserv 12(5):905–920. https://doi.org/10.1023/A:1022869624524
Angienda PO, Lee HJ, Elmer KR, Abila R, Waindi EN, Meyer A (2011) Genetic structure and gene flow in an endangered native tilapia fish (Oreochromis esculentus) compared to invasive Nile tilapia (Oreochromis niloticus) in Yala swamp, East Africa. Conserv Genet 12(1):243–255. https://doi.org/10.1007/s10592-010-0136-2
Blackwell T, Ford AG, Ciezarek AG, Bradbeer SJ, Gracida Juarez CA, Smith AM, Ngatunga BP, Shechonge A, Tamatamah R, Etherington G, Haerty W (2020) Newly discovered cichlid fish biodiversity threatened by hybridization with non-native species. Mol Ecol 30:895–911. https://doi.org/10.1101/2020.08.06.240002
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
ChampneysT, Genner MJ, Ioannou CC (2021) Invasive Nile tilapia dominates a threatened indigenous tilapia in competition over shelter. Hydrobiologia 848(16):3747–3762. https://doi.org/10.1007/s10750-020-04341-8
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134(3):959–969. https://doi.org/10.1093/genetics/134.3.959
Crispo E, Chapman LJ (2008) Population genetic structure across dissolved oxygen regimes in an african cichlid fish. Mol Ecol 17(9):2134–2148. https://doi.org/10.1111/j.1365-294X.2008.03729.x
Curto M, Winter S, Seiter A, Schmid L, Scheicher K, Barthel LM, Plass J, Meimberg H (2019) Application of a SSR-GBS marker system on investigation of european hedgehog species and their hybrid zone dynamics. Ecol Evol 9(5):2814–2832. https://doi.org/10.1002/ece3.4960
D’Amato ME, Esterhuyse MM, Van Der Waal BC, Brink D, Volckaert FA (2007) Hybridization and phylogeography of the Mozambique tilapia Oreochromis mossambicus in southern Africa evidenced by mitochondrial and microsatellite DNA genotyping. Conserv Genet 8:475–488. https://doi.org/10.1007/s10592-006-9186-x
Deines AM, Bbole I, Katongo C, Feder JL, Lodge DM (2014) Hybridisation between native Oreochromis species and introduced Nile tilapia Oreochromis niloticus in the Kafue River. Zambia Afr J Aquat Sci 39(1):23–34. https://doi.org/10.2989/16085914.2013.864965
Diedericks G, Maetens H, Van Steenberge M, Snoeks J (2021) Testing for hybridization between Nile tilapia (Oreochromis niloticus) and blue spotted tilapia (Oreochromis leucostictus) in the Lake Edward system. J Great Lakes Res 47(5):1446–1452. https://doi.org/10.1016/j.jglr.2021.06.005
Eknath AE, Hulata G (2009) Use and exchange of genetic resources of Nile tilapia (Oreochromis niloticus). Rev Aquac 1(3–4):197–213. https://doi.org/10.1111/j.1753-5131.2009.01017.x
Firmat C, Alibert P, Losseau M, Baroiller JF, Schliewen UK (2013) Successive invasion-mediated interspecific hybridizations and population structure in the endangered cichlid Oreochromis mossambicus. PLoSOne 8(5):e63880. https://doi.org/10.1371/journal.pone.0063880
Garnier-Géré P, & Chikhi L (2013) Population subdivision, Hardy–Weinberg equilibrium and the Wahlund effect. eLS.
Gu DE, Mu XD, Song HM, Luo D, Xu M, Luo JR, Hu YC (2014) Genetic diversity of invasive Oreochromis spp.(tilapia) populations in Guangdong province of China using microsatellite markers. Biochem Syst Ecol 55:198–204. https://doi.org/10.1016/j.bse.2014.03.035
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symp. Ser. (Vol. 41, pp. 95–98)
Hickley P, Bailey R, Harper DM, Kundu R, Muchiri M, North R, Taylor A (2002) The status and future of the Lake Naivasha fishery, Kenya. Hydrobiologia 488(1–3):181–190. https://doi.org/10.1023/A:1023334715893
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9(5):1322–1332. https://doi.org/10.1111/j.1755-0998.2009.02591.x
Jiang B, Fu J, Dong Z, Fang M, Zhu W, Wang L (2019) Maternal ancestry analyses of red tilapia strains based on D-loop sequences of seven tilapia populations. PeerJ 7:e7007. https://doi.org/10.7717/peerj.7007
Kajungiro RA, Palaiokostas C, Pinto FAL, Mmochi AJ, Mtolera M, Houston RD, De Koning DJ (2019) Population structure and genetic diversity of Nile tilapia (Oreochromis niloticus) strains cultured in Tanzania. Front Genet 10:1269. https://doi.org/10.3389/fgene.2019.01269
Kariuki J, Tibihika PD, Curto M, Alemayehu E, Winkler G, Meimberg H (2021) Application of microsatellite genotyping by amplicon sequencing for delimitation of african tilapiine species relevant for aquaculture. Aquac 736501. https://doi.org/10.1016/j.aquaculture.2021.736501
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Kwikiriza G, Yegon MJ, Byamugisha N, Beingana A, Atukwatse F, Barekye A, Nattabi JK, Meimberg H (2023) Morphometric variations of Nile Tilapia (Oreochromis niloticus)(Linnaeus, 1758) local strains collected from different fish farms in South Western Highland Agro-Ecological Zone (SWHAEZ), Uganda: screening strains for aquaculture. Fishes 8(4):217. https://doi.org/10.3390/fishes8040217
Labiros DA, Catalig AMP, Ymbong RRJ, Sakuntabhai A, Lluisma AO, Edillo FE (2022) Novel and broadly Applicable microsatellite markers in identified chromosomes of the Philippine Dengue Mosquitoes, Aedes aegypti (Diptera: Culicidae). J Med Entomol 59(2):545–553. https://doi.org/10.1093/jme/tjab194
Langella O (1999) Populations 1.2. 28: a population genetic software. http://www.pge. cnrs-gif. fr/bioinfo/populations/index. php
Laurent C, Yongo E, Waithaka E, Mutethya E (2020) Analysis of some biological aspects of the Blue-spotted tilapia, Oreochromis leucostictus (Trewavas, 1933) in Lake Naivasha, Kenya. https://doi.org/10.31219/osf.io/6xf7j
Leigh JW, Bryant D (2015) popart: full-feature software for haplotype network construction. Methods Ecol Evol 6(9):1110–1116. https://doi.org/10.1111/2041-210X.12410
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Martinez AS, Willoughby JR, Christie MR (2018) Genetic diversity in fishes is influenced by habitat type and life-history variation. Ecol Evol 8(23):12022–12031. https://doi.org/10.1002/ece3.4661
Matsuishi T, Muhoozi L, Mkumbo O, Budeba, Njiru M, Asila A, Othina A, Cowx IG (2006) Are the exploitation pressures on the Nile perch fisheries resources of Lake Victoria a cause for concern? Fish Manag Ecol 13(1):53–71. https://doi.org/10.1111/j.1365-2400.2006.00477.x
Meyer BS, Matschiner M, Salzburger W (2015) A tribal level phylogeny of Lake Tanganyika cichlid fishes based on a genomic multi-marker approach. Mol Phylogenet Evol 83:56–71. https://doi.org/10.1016/j.ympev.2014.10.009
Munguti JM, Kim JD, Ogello EO (2014) An overview of kenyan aquaculture: Status, challenges, and opportunities for future development. Fish Aquat Sci 17(1):1–11. https://doi.org/10.5657/FAS.2014.0001
Mwanja W, Booton GC, Kaufman L, Chandler M, Fuerst P, Donaldson E, & MacKinlay D (1996) Population and stock characterization of Lake Victoria. Tilapine fishes based on RAPD markers. In Aqyuaculture Biotechnology Symposium Prodeedings of the International Congress on the Biology of Fishes (ed, by EM Donaldson & DD Mackinlay) (pp. 115–124).
Mwanja WW, Kaufman L, Fuerst PA (2010) Comparison of the genetic and ecological diversity of the native to the introduced tilapiines (Pisces: Cichlidae), and their population structures in the Lake Victoria region, East Africa. Aquat Ecosyst Health Manag 13(4):442–450. https://doi.org/10.1080/14634988.2010.527268
Ndiwa TC, Nyingi DW, Agnese JF (2014) An important natural genetic resource of Oreochromis niloticus (Linnaeus, 1758) threatened by aquaculture activities in Loboi drainage, Kenya. PLoS ONE 9(9):e106972. https://doi.org/10.1371/journalpone0106972
Njiru M, Mkumbo OC, Van der Knaap M (2010) Some possible factors leading to decline in fish species in Lake Victoria. Aquat Ecosyst Health Manag 13(1):3–10. https://doi.org/10.1080/14634980903566253
Ogutu-Ohwayo R, Hecky RE (1991) Fish introductions in Africa and some of their implications. Can J Fish Aquat Sci 48(S1):8–12. https://doi.org/10.1139/f91-299
Okwiri B, Donde OO, Kibet CJ (2019) Status and impacts of non-native freshwater fish on fisheries biodiversity and biogeography in Kenya: a management perspective. Lake Reserv Manag 24(4):332–343. https://doi.org/10.1111/lre.12291
Opiyo MA, Marijani E, Muendo P, Odede R, Leschen W, Charo-Karisa H (2018) A review of aquaculture production and health management practices of farmed fish in Kenya. Int J Vet Sci Med 6(2):141–148. https://doi.org/10.1016/j.ijvsm.2018.07.001
Parakatselaki ME, Ladoukakis ED (2021) mtDNA heteroplasmy: origin, detection, significance, and Evolutionary Consequences. Life 11(7):633. https://doi.org/10.3390/life11070633
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Salzburger W, Meyer A (2004) The species flocks of east african cichlid fishes: recent advances in molecular phylogenetics and population genetics. Sci Nat 91(6):277–290. https://doi.org/10.1007/s00114-004-0528-6
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver. 2.000. A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
Shechonge A, Ngatunga BP, Tamatamah R, Bradbeer SJ, Harrington J, Ford AGP, Turner GF, Genner MJ (2018) Losing cichlid fish biodiversity: genetic and morphological homogenization of tilapia following colonization by introduced species. Conserv Genet 19:1199–1209. https://doi.org/10.1007/s10592-018-1088-1
The State of World Fisheries and Aquaculture (2022) FAO. https://doi.org/10.4060/cc0461en
Tibihika PD, Curto M, Alemayehu E, Waidbacher H, Masembe C, Akoll P, Meimberg H (2020) Molecular genetic diversity and differentiation of Nile tilapia (Oreochromis niloticus, L. 1758) in east african natural and stocked populations. BMC Evol Biol 20(1):16. https://doi.org/10.1186/s12862-020-1583-0
Tibihika PD, Curto M, Dornstauder-Schrammel E, Winter S, Alemayehu E, Waidbacher H, Meimberg H (2019) Application of microsatellite genotyping by sequencing (SSR-GBS) to measure genetic diversity of the East African Oreochromis niloticus. Conserv Genet 20(2):357–372. https://doi.org/10.1007/s10592-018-1136-x
Acknowledgements
We kindly acknowledge Austrian Development Corporation (ADC) for funding this work. Special thanks go to International Training Programmes in Limnology (IPGL) for the support towards the write up of this paper. We would like to acknowledge fisheries officers managing the different beaches where field work took place for their contribution towards sample collection. We are also grateful to the beach management unit (BMU) for their kind cooperation during sampling.
Funding
Open access funding provided by University of Natural Resources and Life Sciences Vienna (BOKU).
Author information
Authors and Affiliations
Contributions
Harald Meimberg, Gerold Winkler and Gerald Kwikiriza conceptualized the idea, sample collection was done by Gerald Kwikiriza and John Kariuki, sample processing and genotyping was carried out by Gerald Kwikiriza and Thapasya Vijayan. Gerald Kwikiriza, Juliet Kigogo Nattabi, Manuel Curto and Papius Dias Tibihika analyzed the data. Funding acquisition was by Gerold Winkler. All the authors participated in manuscript preparation, review and editing. All authors have read and agreed to the submitted version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of Interest
The authors declare no conflicting interests in this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kwikiriza, G., Vijayan, T., Tibihika, P.D. et al. Introgressive hybridization levels of Tilapiine species in Lake Victoria basin, Kenya inferred from microsatellite and mitochondrial DNA genotyping based on next-generation sequencing. Conserv Genet 25, 305–318 (2024). https://doi.org/10.1007/s10592-023-01570-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-023-01570-x