Abstract
When backcrosses are fertile, interbreeding between endangered taxa can lead to the admixture of gene pools under threat. One such case pertains to the Mesoamerican crocodile Crocodylus moreletii, a species which shows strong signatures of both recent hybridisation and historic intogression with the American crocodile C. acutus across large parts of its range. In the present paper, we use RAD-seq derived SNPs (4980 nuclear and seven mtDNA loci) to demonstrate that C. moreletii populations inhabiting the region of Calakmul in central Yucatan (Mexico) are rather unaffected by hybridization, despite being surrounded by coastal areas where pervasive admixture has previously been documented. All (based on fastSTRUCTURE) and 96% (based on NGSadmix) of 84 genotyped individuals from 18 sampled waterbodies (locally termed aguadas) were free from nuclear introgression of C. acutus DNA at at threshold of 0.95. Seven individuals (8%) possessed a C. acutus mtDNA haplotype, five of which were derived from two adjacent, rather peripheral aguadas. Spatial inferences based on a DAPC and fineRADstructure further showed that the region of Calakmul is inhabited by three genetic clusters spanning across a set of distinct aguadas each. Taken together, our findings reveal that central Yucatan contains the currently largest documented stronghold of C. moreletii populations only marginally affected by introgression, which has major implications for the conservation management of this important flagship species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Knowledge of the amount and distribution of genetic variation across the range of given species facilitates the designation of conservation units, and aids in the identification of threats such as inbreeding depression and loss of evolutionary potential (e.g. Frankham et al. 2017; Allendorf et al. 2022). Boundaries between species can however also be blurred by hybridisation and introgression, leading to situations where conservation frameworks which are based on the biological species concept become difficult to apply (e.g. Allendorf et al. 2001; Fitzpatrick et al. 2015; vonHoldt et al. 2018). Fuelled by the straightforward identification of admixed individuals through genomic data and the fact that human perturbations to natural systems increase the opportunity for species to interbreed, evidence has recently mounted that incomplete reproductive barriers between species are more common than traditionally assumed (Grabenstein and Taylor 2018; Taylor and Larson 2019). Introgression is further a major conservation concern per se, as it can cause extinction through genetic swamping (Rhymer and Simberloff 1996; Todesco et al. 2016).
Crocodylians represent a morphologically conserved group of top predators comprising 24 recognized species (Grigg and Kirshner 2015), with genetic data however currently challenging a range of traditionally assumed biogeographies and taxonomic relationships (Meredith et al. 2011; Pacheco-Sierra et al. 2016; Muniz et al. 2018; Roberto et al. 2020; Avila-Cervantes et al. 2021; Zucoloto et al. 2021). While hybridisation is recognised both as a force of diversification as well as a threat to endangered crocodylian taxa (Fitzsimmons et al. 2002; Milian-Garcia et al. 2016; Nguyen et al. 2018; Pacheco-Sierra et al. 2018; Chattopadhyay et al. 2019; Pacheco-Sierra and Amavet 2021), information on its incidence, geographical extent and driving factors among wild populations is still limited.
The Morelet’s crocodile (Crocodylus moreletii) is a medium-to-large species occurring in Atlantic regions of predominately Mexico, with smaller occurences in Guatemala and Belize. A large part of the C. moreletii range is situated in parapatry with the American crocodile C. acutus, and hybridisation between the two species has initially been suggested based on morphological evidence (Ross and Ross 1974; Ross and Mayer 1983). Local interbreeding between C. moreletii and C. acutus has subsequently been regularly reported based on mtDNA sequence data and microsatellites (Ray et al. 2004; González-Trujillo et al. 2012; Hekkala et al. 2015; Serrano-Gomez et al. 2016), also revealing that hybrids cannot be unambiguously identified based on phenotypes alone (Cedeño-Vázquez et al. 2008). Remarkably, recent evidence based on high-throughput DNA data further suggests that a large number of populations routinely identified as C. moreletii actually consist of individuals which are admixed, giving rise to conservation concerns and questioning the general status of the species itself (Pacheco-Sierra et al. 2016, 2018).
One example of widespread co-occurrence between C. moreletii and C. acutus pertains to the Yucatan Peninsula, which separates the Golf of Mexico from the Caribbean Ocean. While adjacent islands such as Cozumel are exclusively inhabited by C. acutus, wide areas along the Yucatan coast are occupied by hybrid populations of varying levels of admixture (e.g. Cedeño-Vázquez et al. 2008; Hekkala et al. 2015; Pacheco-Sierra et al. 2016, 2018; Soria-Ortiz et al. 2022). The centre of the peninsula includes the Calakmul Biosphere Reserve (CBR), which is characterised by higher elevation than its surroundings (approx. 250 m a.s.l.) and geological characteristics which lead to the formation of temporary or permanent waterbodies sustained by rainfall (locally termed aguadas, e.g. García-Gil 2000; Gunn et al. 2002; Barão-Nóbrega 2019). Remarkably, these aguadas have recently been shown to harbour ~ 10,000 putative C. moreletii individuals (about 7–13% of the total population estimated for Mexico; Barão-Nóbrega et al. 2022), although their genetic integrity remains to be investigated. In the present paper, we use ddRAD-derived nuclear and mitochondrial SNPs to (i) screen C. moreletii individuals residing in the Calakmul region for evidence of hybridization and introgression with C. acutus, and (ii) document the local population genetic structure of C. moreletii across the sampled aguadas.
Material and methods
Fieldwork and sample collection
Fieldwork was conducted between 2017 and 2018 at 18 waterbodies across six sampling areas within the region of Calakmul (denoted CK1-6, Fig. 1). In total, 85 crocodiles were captured at night with a pole with a break-away noose and restrained using a pole-snare (Ketch-All Animal Restraining Pole), ropes and tapes (following Sánchez-Herrera et al. 2011). Individuals were marked by removal of an individual combination of up to three vertical tail scutes (Sánchez-Herrera et al. 2011; Barão-Nóbrega et al. 2016), stored in 96% ethanol as a source for DNA. All procedures were performed on site, and individuals were released within a maximum period of 30 min after capture. In addition to the individuals sampled within and around the CBR, four C. acutus reference tissue samples from Banco Chinchorro and Cozumel islands in Quintana Roo (Pacheco-Sierra et al. 2016, 2018), and five C. moreletii reference samples based on rescued crocodiles captured in urbanised areas in Altamira (Tamaulipas) about 1000 km north of Calakmul were included in the analysis (Figure S1). All field activities were performed in compliance with the guidelines for use of live amphibians and reptiles in field and laboratory research (Beaupre et al. 2004), and under a research permit issued by the relevant Mexican authorities (SGPA/DGVS/004761/18).
Elevation profile of the region of Calakmul in the southern-central portion of the Yucatan Peninsula (Mexico). The area inside the black line represents the Calakmul Biosphere Reserve. White circles represent the 18 waterbodies where samples of Crocodylus moreletii were obtained. Letters identify each of the six sampling areas within region of Calakmul. The elevation data shown in this map was obtained from a spaceborne digital elevation model (DEM) provided by MERIT DEM (Yamazaki et al. 2017)
Genotyping
DNA extraction was conducted from approximately 20 mg of tissue at the Aquatic Ecology and Systematics Department of El Colegio de la Frontera Sur (ECOSUR), Chetumal, Mexico, using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. DNA concentration was assessed using a microvolume spectrophotometer (Eppendorf™) and normalised to approximately 20 ng/μL. To verify standardised concentration and to ensure that the extracted DNA was of high molecular weight, samples were visualised on a 1.5% agarose gel. Preparation of dd-RAD libraries and DNA sequencing were performed by Floragenex following the protocol of Truong et al. (2012). In brief, DNA was double digested with a combination of rare and frequent cutting endonucleases (PstI and MseI, respectively), followed by ligation with adaptors with individual indices. 1 × 100 bp single-end sequencing was performed on the resulting PCR-generated library using an Illumina HiSeq 4000.
Returned raw sequences were filtered, demultiplexed and processed using the software STACKS 2.0 (Catchen et al. 2013; Rochette et al. 2019), following existing protocols (Rochette and Catchen 2017). Sequences were checked for correct restriction sites and adaptor sequences using the default values in the process_radtags pipeline. Reads with an uncalled base were discarded, as were reads containing a 15 bp window in which the average quality dropped below a Phred score of 10 (i.e. a 90% probability of being correct). Barcodes and RAD-tags containing up to one mismatch to an expected sequence were retained.
In the absence of reference genomes for C. moreletii or C. acutus, the genome of C. porosus (Green et al. 2014) was obtained from Ensembl (https://www.ensembl.org/Crocodylus_porosus/Info/Index) for single nucleotide polymorphism (SNP) discovery and genotyping also in STACKS. A reference-aligned assembly was produced by mapping the clean demultiplexed sequences against the C. porosus genome using the software GSNAP (Wu et al. 2016), analysed using the ref_map pipeline (Rochette et al. 2019) following procedures described in Paris et al. (2017) and Rochette and Catchen (2017). In brief, the ref_map pipeline was run with varying parameter values while recording the number of polymorphic loci found across at least 80% of samples (the r80 loci, Paris et al. 2017; Rochette and Catchen 2017) to identify a stable set of values at a mean final coverage of 25.6x (standard deviation: 4.4x). Prior to exporting the genotype calls from STACKS for downstream analyses, the dataset was further filtered using the populations pipeline. In addition to only retaining the r80 loci, loci with a minor allele frequency below 0.05, an observed heterozygosity above 0.70, and loci absent in at least one of the three sample types (C. acutus and C. moreletii reference samples, and C. moreletii from Calakmul) were removed.
Mitochondrial sequences were identified by separately mapping the clean demultiplexed reads against both the mitochondrial genomes of C. acutus (NCBI Reference Sequence: NC_015647.1; Man et al. 2011) and C. moreletii (NCBI Reference Sequence: NC_015235.1; Meganathan et al. 2011). Mapped reads were sorted and indexed using SAMTOOLS v.1.4 (Li et al. 2009). The software IGV (Robinson et al. 2011) was used to visually assess where the reads map against the mitochondrial genomes of both species (following Barth et al. 2020), and four regions were identified (2278–2872, 5836–5930, 8392–8485 and 9528–9622); as these regions were detected in both species, we continued using reads mapped against C. acutus only. Sequences from all samples mapped to these regions were extracted and converted to FASTA format using SAMTOOLS, BCFTOOLS v.1.6 (Li 2011), and SEQTK v.1.0 (https://github.com/lh3/seqtk).
Data analysis
Observed (Ho) and expected (He) heterozygosities across genotyped individuals were calculated using the R package ADEGENET (Jombart 2008; Jombart and Ahmed 2011), supplemented with information on total gene diversity (Ht) and genetic diversity within sampling areas (Hs) calculated using the R package MMOD (Winter 2012). The level of nuclear admixture across all samples was examined using two complementary approaches: a Bayesian inference as implemented in fastSTRUCTURE (Raj et al. 2014), and a maximum likelihood approach implemented in the software NGSadmix (Skotte et al. 2013). Both methods identify individual admixture proportions, with fastSTRUCTURE using SNP calls and NGSadmix using genotype likelihood estimates produced through the software ANGSD (Korneliussen et al. 2014). To quantify levels of hybridization between C. moreletii and C. acutus, the estimated admixture proportions at two assumed genetic clusters representing the species (K = 2) were used with default convergence priors and five replicates for each value. Individuals exhibiting point estimates of qi > 0.95 with the lower bound of the 95% credible intervals (CI) for qi > 0.8 were assumed as non-admixed (following Rodriguez et al. 2008; Pacheco-Sierra et al. 2016); a qi threshold of 0.99 was also considered for comparison. For NGSadmix, runs at K ranging from 2 to 10 were additionally performed to determine the optimal value of K across the considered populations using CLUMPAK (Kopelman et al. 2015).
For the analyses of mitochondrial sequences, the four identified regions were aligned with default settings in MAFFT v.7.397 (Katoh and Standley 2013), and the software TASSEL v.5.0 (Bradbury et al. 2007) was used to visually assess the mapped sequences across all samples. All four mapped regions were further aligned against other published mitochondrial isolates of C. acutus (GenBank References: JF502241.1; JF315769.1, JF315757.1, JF315747.1, JF315714.1, HM636894.1, JF315729.1; Meredith et al. 2011; Oaks 2011) and C. moreletii (GenBank References: JF315768.1, JF315752.1; HQ585889.1; Meganathan et al. 2011; Oaks 2011) using the NCBI database BLAST tool (Zhang et al. 2000; Morgulis et al. 2008). Two regions (5836–5930 and 8392–8485) were discarded due to being undiagnostic at the level of species. The other two regions (2278–2372 and 9528–9622) were retained, and used for all analyses.
The spatial genetic structure within Calakmul was further assessed using a Discriminant Analysis of Principal Components (DAPC; Jombart et al. 2010) as implemented in the ADEGENET package (Jombart 2008; Jombart and Ahmed 2011) in RStudio version 1.1.456 (RStudio Team 2020). DAPCs seek to maximise between-group variation whilst minimising variation within groups, and combined with sample locality information reveal the spatial partitioning of genetic variation. Total gene diversity (Ht) and genetic diversity (Hs) within clusters identified by the DAPC were again derived using MMOD (Winter 2012). Pairwise Fst values between derived clusters were calculated using the R package HIERFSTAT (Goudet 2005).
The software fineRADstructure v.0.3.1 (Malinsky et al. 2018) was additionally used to infer shared ancestry among individuals by clustering them according to similarity of their RAD haplotypes. This approach exploits information drawn from stacks containing several SNPs, whereas different stacks are assumed to be unlinked to derive a co-ancestry matrix based on the most recent coalescent events (i.e. the sharing of identical or nearest-neighbour haplotypes among individuals). Haplotype data were converted for fineRADstructure input using the script Stacks2fineRAD.py (Malinsky et al. 2018). The co-ancestry matrix was inferred using RADpainter, setting the number of MCMC burn-in iterations to 100,000, the sample iterations to 100,000, and the thinning interval to 1,000 (Lawson et al. 2012). To reflect the relationships within the co-ancestry matrix, the inferred clusters were arranged according to a tree inferred with fineRADstructure using 100,000 hill-climbing iterations, allowing for all possible tree comparisons (following Barth et al. 2020).
Results
Two samples from Calakmul were discarded due to a low number of reads (~ 5200 and 70,000, respectively). Across the remaining 93 samples, the number of reads per individual retained after filtering ranged between 2.31*106 and 5.42*106 (mean: 3.07*106) and resulted in a final panel of 4980 polymorphic SNPs. Average heterozygosity across the 84 C. moreletii genotyped from the region of Calakmul was 0.273 (standard error: 0.001). Average heterozygosities amongst the five reference samples of C. moreletii and four reference samples of C. acutus were 0.182 (standard error = 0.003) and 0.100 (se: 0.003), respectively. Total gene diversity (Ht) and genetic diversity of C. moreletii within the six sampling areas in Calakmul (Hs) was 0.32 and 0.26, respectively.
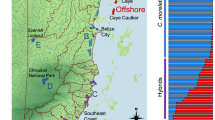
Bayesian admixture proportions (qi) for K = 2 estimated in fastSTRUCTURE identified 100% (84/84) and 86% (72/84) of crocodile samples from the region of Calakmul as non-admixed C. moreletii at qi thresholds of 0.95 and 0.99, respectively (Fig. 2). The C. acutus reference samples from Cozumel and Banco Chinchorro were free from admixture, whereas substantial admixture proportions were observed in all non-Calakmul C. moreletii reference samples (0.17 ± 0.01; 0.10–0.18). Similarly, admixture proportions estimated from genotype likelihoods for K = 2 in NGSadmix identified 96% (81/84) and 74% (62/84) of C. moreletii samples from Calakmul as non-admixed at thresholds of 0.95 and 0.99 (Fig. 2), respectively. Again, substantial admixture proportions (0.18 ± 0.04; 0.11–0.23) were apparent in all C. moreletii reference samples.
Genetic structure at K = 2 using Bayesian admixture proportions (qi) from fastSTRUCTURE (top panel, Raj et al. 2014) and ancestry proportions (Q-scores) from NGSadmix (bottom panel, Skotte et al. 2013), where solid lines are 95% credible intervals and different colours represent each admixture proportion. Each vertical bar corresponds to one crocodile sample. Inverted black triangles represent samples with Crocodylus acutus mitochondrial haplotypes. Dashed white lines delimit different sampling sites within each of the six general areas surveyed within the region of Calakmul (CK). CA and CM represent, respectively, the external reference samples of the C. acutus and C. moreletii
The mapped mtDNA reads revealed six species-diagnostic SNPs, with all reference samples possessing the species-specific haplotype (positions 2328, 2367, 9543, 9570, 9582, 9618, Figure S2). Seventy-seven (92%) crocodiles from Calakmul exhibited the C. moreletii haplotype, whilst the remaining seven crocodiles exhibited the C. acutus haplotype (Fig. 2).
The CLUMPAK method suggested that K = 4 represented the best fit to the population partitions as inferred by NGSadmix across the entire dataset, confirming genetic sub-structuring also within Calakmul (one cluster of C. acutus and three clusters of C. moreletii, including the reference samples which were part of one cluster; Figure S3). Accordingly, the DAPC confirmed a distinct structuring of C. moreletii within the region of Calakmul into three main clusters (Fig. 3), corresponding to western, central and eastern regions. Apart from five individuals, all C. moreletii sampled from a given sampling area were assigned to the same cluster (Fig. 3). The co‐ancestry matrix resulting from the fineRADstructure analysis further confirmed the partitioning into three clusters for the CBR, with identical assignments of individuals to given clusters as in the DAPC (Fig. 4). Total gene diversity (Ht) and genetic diversity within identified clusters (Hs) was 0.32 and 0.28, respectively. Pairwise Fst values between clusters were 0.13 between the western and eastern clusters of Calakmul, and 0.18 between both of these and the central cluster (Fig. 3).
Spatial distribution of Crocodylus moreletii samples from the three genetic clusters inferred by the k means algorithm in the Discriminant Analysis of Principal Components (DAPC; Jombart et al. 2010). Circles containing the letter A indicate samples with reads matching regions 2278–2872 and 9528–9622 of the Crocodylus acutus mitochondrial genome. The numbered black circles indicate the sample size in that general location. The figure on the bottom left corner is the DAPC plot generated from the SNP dataset of C. moreletii from Calakmul. The first two PC axes indicate differentiation across geographical sample regions
Individual co-ancestry based on haplotype similarity across all samples of C. moreletii and C. acutus (top panel), and C. moreletii samples from Calakmul (bottom panel), estimated with fineRADstructure (Malinsky et al. 2018). Heatmap colours indicate numbers of RAD loci with estimated shared co-ancestry. Individuals are listed on both axes in the same order, clustered according to the tree shown on top of the heatmap (Lawson et al. 2012)
Discussion
Hybridization and introgression are an integral part of the evolutionary history of many species (e.g. Mallet 2005; Abbott et al. 2013; Payseur and Rieseberg 2016). In a conservation context, however, hybridisation also represents an increasing concern: admixed individuals are difficult to capture in legislative frameworks, and environmental perturbation as well as the spread of non-native species can lead to the interbreeding of taxa which would otherwise not meet (Allendorf et al. 2001; Grabenstein and Taylor 2018; vonHoldt et al. 2018). The present study focuses on the crocodile C. moreletii, a species which is characterised by pervasive admixture with the congener C. acutus, and demonstrates that central Yucatan serves as an important stronghold for populations which are only marginally affected by introgression. The study further demonstrates that the study area is divided into three distinct genetic clusters spanning across a range of hydrologically dynamic sites each. Below, we discuss these findings in view of conservation considerations for this important umbrella crocodylian species.
When quantifying individual levels of hybridization and introgression, a set of non-admixed reference samples from both parental species usually form the basis for the estimation of admixture proportions for the focal individuals under study (e.g. Barth et al. 2020; Arntzen et al. 2021). Because the sampling for the present study was centred in Calakmul, only limited numbers of reference individuals from outside the study area were however available, with multiple attempts to obtain high-quality DNA from other reference samples having been unsuccessful (data not shown). Despite our analyses revealing that the C. moreletii reference samples from Altamira had higher levels of nuclear introgression than Calakmul itself (see also Pacheco-Sierra et al. 2016, 2018), the ~ 5000 nuclear SNPs combined with the RAD-derived mtDNA sequence information yielded converging findings across differential analytical pathways, and we therefore deemed our conclusions sufficiently robust.
Our analyses suggest that a substantial proportion of C. moreletii individuals from Calakmul are virtually free from C. acutus introgression, and all (based on fastSTRUCTURE) or 96% (based on NGSadmix) of individuals were below an admixture value of 0.05, a common threshold to define non-admixed individuals in hybridisation studies (for C. moreletii see Rodriguez et al. 2008; Rodriguez et al. 2008; Hekkala et al. 2015; Pacheco-Sierra et al. 2016). Moreover, all 84 studied individuals had an admixture value below 0.1, suggesting that interspecific matings did not take place for at least four generations. Crocodiles disperse mainly through hydrological networks (coastlines, lagoons, and rivers; Lee 2000; Villela et al. 2008; Velo-Antón et al. 2014; Grigg and Kirshner 2015), and the low levels of admixture of Calakmul populations is therefore likely a result of ecological isolation through an elevated topographic profile, combined with poor hydrological connectivity to surrounding areas (García-Gil 2000; Gunn et al. 2002; see also Fig. 1). While hybridisation in coastal areas of Yucatan is assumed to date back 2–3 million years (Pacheco-Sierra et al. 2018), our current analyses are unable to discern whether the low levels of introgressed alleles observed for some C. moreletii individuals in Calakmul for example stem from the colonisation of the area by already admixed individuals, or from occasional interspecific gene flow across environmental gradients.
Likely linked to low metabolic rates, high longevity as well as low mutation rates during DNA replication, crocodylians generally exhibit low levels of mtDNA variation (Glenn et al. 2002; Rodriguez et al. 2008; Galtier et al. 2009; for examples on C. moreletii see; González-Trujillo et al. 2012), and we indeed did not find any intraspecific variation among the retrieved mtDNA sequences. Despite levels of nuclear introgression which were not indicative of recent hybridisation, seven out of the 84 individuals from Calakmul were characterised by bearing a C. acutus mtDNA haplotype. Five out of these seven individuals were confined to two adjacent aguadas (from where nine samples were available, i.e. more than half of all individuals possessed disconcordant mtDNA), supporting the existence of a marked population structure within the CBR (see also below). It is also worth noting that six out of seven individuals with C. acutus mtDNA haplotypes were assigned to the same genetic cluster, and that discordant haplotypes were only found in areas of rather low altitude (the eastern and western sides of the Xbonil Hills, Sierrita de Ticul; Fig. 1). We are however presently unable to determine whether the observed cases of nuclear-mitochondrial discordance are linked to incomplete lineage sorting during divergence or ancient hybridisation (see Toews and Brelsford 2012; Bonnet et al. 2017 for general discussions).
The present study revealed the existence of three spatial genetic clusters across the 18 sampled waterbodies, suggesting high levels of connectivity between some waterbodies but also restricted gene flow across parts of Calakmul. Considerable genetic differentiation between geographical regions with poor hydrological connectivity despite high gene flow and absence of genetic structure within a geographical region has also been previously reported in other crocodylian species (e.g. Crocodylus suchus—Velo-Antón et al. 2014; Paleosuchus trigonatus—Muniz et al. 2019; Crocodylus johnstoni—Cao et al. 2020). While permanent aquatic dispersal corridors such as rivers are absent, Calakmul is characterised by temporary widespread flooding of lowland forests during particularly strong rainy seasons (García-Gil 2000; Gunn et al. 2002; Márdero et al. 2019), which is likely responsible for the observed spatial genetic partitioning of the study area. Two individuals (one sub-adult and one adult male) assigned to Cluster 3 were found in the area of Cluster 1, and three individuals (all female sub-adults) assigned to Cluster 1 were found in the area of Cluster 2 (Fig. 3). Disregarding potential sampling errors, the lack of individuals which are genetically intermediate between clusters suggests that such potential migrants do not reproduce outside their natal areas. Further individual-based analyses of the obtained genotypes are planned to be published in a forthcoming paper focusing on relatedness structures, inferred parentages and mating systems across the sampled aguadas (Barão-Nóbrega et al., in preparation).
What do our findings mean for the conservation management of C. moreletii? Although hybridization between wild crocodiles has long been documented, its importance for conservation has so far received rather little attention (e.g. Hekkala 2004; Zucoloto et al. 2021). In the case of C. moreletii, concerns have recently been raised that the species predominately constitutes several allopatric C. moreletii—C. acutus hybrid lineages, with the only previously known non-admixed C. moreletii populations confined to upstream continental lagoons in Northern Mexico (Pacheco-Sierra et al. 2016, 2018). From this view, the findings from the present study that the region of Calakmul supports further populations which are largely free from introgression is of major conservation importance, particularly since the region harbours approximately 10% of the Mexican population of C. moreletii (~ 10,000 individuals, Barão-Nóbrega et al. 2022). The long-term existence of large populations within Calakmul is also supported by levels of genetic variation which appear higher than in the previously known populations which are free from widespread admixture (Ho = 0.19, compared to Ho = 0.13 in northern Mexico, see Pacheco-Sierra et al. 2018; both values are based on RAD-derived SNPs).
Crocodylus moreletii is currently protected by Mexican law as conservation dependant (NOM-059-ECOL-2001) and internationally listed by CITES under Appendix II. However, existing conservation guidelines and legislations are regarded as in need of review, due to the extent and distribution of hybrid lineages and because non-admixed populations require different categorization than their hybrid counterparts (Pacheco-Sierra et al. 2016, 2018). Regarding the future safeguarding of populations within Calakmul, care should be devoted to preventing possible inward dispersal by C. moreletii—C. acutus for example facilitated by future infrastructure projects, also avoiding translocations and release of captive individuals. Future studies should aim to increase our understanding of the link between the differential habitat requirements of C. moreletii and C. acutus with their potential to interbreed, in order to build a predictive framework of levels of hybridization across the species ranges.
References
Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJ, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Most M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Vainola R, Wolf JB, Zinner D (2013) Hybridization and speciation. J Evol Biol 26:229–246. https://doi.org/10.1111/j.1420-9101.2012.02599.x
Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001) The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16:613–622. https://doi.org/10.1016/S0169-5347(01)02290-X
Allendorf FW, Funk WC, Aitken SN, Byrne M, Luikart G (2022) Conservation and the genomics of populations. Oxford University Press, Oxford
Arntzen JW, Jehle R, Wielstra B (2021) Genetic and morphological data demonstrate hybridization and backcrossing in a pair of salamanders at the far end of the speciation continuum. Evol Appl 14:2784–2793. https://doi.org/10.1111/eva.13312
Avila-Cervantes J, Arias C, Venegas-Anaya M, Vargas M, Larsson HCE, McMillan WO (2021) Effect of the Central American Isthmus on gene flow and divergence of the American crocodile (Crocodylus acutus). Evolution 75:245–259. https://doi.org/10.1111/evo.14139
Barão-Nóbrega JAL (2019) Aguadas of Calakmul: An update on location and general structure information of waterbodies in the region of Calakmul, Campeche, Mexico. Database published on ResearchGate
Barão-Nóbrega JAL, Puls S, Acton C, Slater K (2016) Crocodylus moreletii (Morelet’s crocodile). Movement. Herpetol Rev 47:291–291
Barão-Nóbrega JAL, González-Jaurégui M, Jehle R (2022) N-mixture models provide informative crocodile (Crocodylus moreletii) abundance estimates in dynamic environments. PeerJ 10:e12906. https://doi.org/10.7717/peerj.12906
Barth JMI, Gubili C, Matschiner M, Tørresen OK, Watanabe S, Egger B, Han Y-S, Feunteun E, Sommaruga R, Jehle R, Schabetsberger R (2020) Stable species boundaries despite ten million years of hybridization in tropical eels. Nat Commun 11:1433. https://doi.org/10.1038/s41467-020-15099-x
Beaupre SJ, Jacobson ER, Lillywhite HB, Zamudio K (2004) Guidelines for the use of live amphibians and reptiles in field and laboratory research. American Society of Ichthyologists and Herpetologists, Miami
Bonnet T, Leblois R, Rousset F, Crochet PA (2017) A reassessment of explanations for discordant introgressions of mitochondrial and nuclear genomes. Evolution 71:2140–2158. https://doi.org/10.1111/evo.13296
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Cao R, Somaweera R, Brittain K, FitzSimmons NN, Georges A, Gongora J (2020) Genetic structure and diversity of Australian freshwater crocodiles (Crocodylus johnstoni) from the Kimberley, Western Australia. Conserv Genet 21:421–429. https://doi.org/10.1007/s10592-020-01259-5
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140. https://doi.org/10.1111/mec.12354
Cedeño-Vázquez JR, Rodriguez D, Calme S, Ross JP, Densmore LD III, Thorbjarnarson JB (2008) Hybridization between Crocodylus acutus and Crocodylus moreletii in the Yucatan Peninsula: evidence from mitochondrial DNA and morphology. J Exp Zool Part A 309A:661–673. https://doi.org/10.1002/jez.473
Chattopadhyay B, Garg KM, Soo YJ, Low GW, Frechette JL, Rheindt FE (2019) Conservation genomics in the fight to help the recovery of the critically endangered Siamese crocodile Crocodylus siamensis. Mol Ecol 28:936–950. https://doi.org/10.1111/mec.15023
Fitzpatrick BM, Ryan ME, Johnson JR, Corush J, Carter ET (2015) Hybridization and the species problem in conservation. Curr Zool 61:206–216. https://doi.org/10.1093/czoolo/61.1.206
Fitzsimmons NN, Buchan JC, Lam PV, Polet G, Hung TT, Thang NQ, Gratten J (2002) Identification of purebred Crocodylus siamensis for reintroduction in Vietnam. J Exp Zool 294:373–381. https://doi.org/10.1002/jez.10201
Frankham R, Ballou JD, Ralls K, Eldridge MDB, Dudash MR, Fenster CB, Lacy RC, Sunnucks P (2017) Genetic management of fragmented animal and plant populations. Oxford University Press, Oxford
Galtier N, Jobson RW, Nabholz B, Glémin S, Blier PU (2009) Mitochondrial whims: metabolic rate, longevity and the rate of molecular evolution. Biol Lett 5:413–416. https://doi.org/10.1098/rsbl.2008.0662
García-Gil G (2000) Cuerpos de agua de la Reserva de la Biosfera Calakmul, Campeche. Escala 1:50000 [Waterbodies of Calakmul Biosphere Reserve, Campeche Scale 1:50000]. In: Uso actual de suelo y estado de conservación de la Reserva de la Biosfera Calakmul, Campeche, pp. El Colegio de la Frontera Sur (ECOSUR), Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO)
Glenn TC, Staton JL, Vu AT, Davis LM, Bremer JRA, Rhodes WE, Brisbin IL Jr, Sawyer RH (2002) Low mitochondrial DNA variation among American alligators and a novel non-coding region in crocodilians. J Exp Zool 294:312–324. https://doi.org/10.1002/jez.10206
González-Trujillo R, Rodriguez D, González-Romero A, Forstner MR, Densmore LD III, Reynoso VH (2012) Testing for hybridization and assessing genetic diversity in Morelet’s crocodile (Crocodylus moreletii) populations from central Veracruz. Conserv Genet 13:1677–1683. https://doi.org/10.1007/s10592-012-0406-2
Goudet J (2005) hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186. https://doi.org/10.1111/j.1471-8286.2004.00828.x
Grabenstein KC, Taylor SA (2018) Breaking barriers: Causes, consequences, and experimental utility of human-mediated hybridization. Trends Ecol Evol 33:198–212. https://doi.org/10.1016/j.tree.2017.12.008
Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G, Vandewege MW, John JAS, Capella-Gutiérrez S, Castoe TA (2014) Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346:1254449. https://doi.org/10.1126/science.1254449
Grigg G, Kirshner D (2015) Biology and evolution of crocodylians. Csiro Publishing, Australia
Gunn JD, Foss JE, Folan WJ, Carrasco MdRD, Faust BB (2002) Bajo sediments and the hydraulic system of Calakmul, Campeche, Mexico. Anc Mesoam 13:297–315. https://doi.org/10.1017/S0956536102132184
Hekkala ER (2004) Conservation genetics at the species boundary: case studies from African and Caribbean crocodiles (Genus: Crocodylus). Columbia University New York, New York
Hekkala ER, Platt SG, Thorbjarnarson JB, Rainwater TR, Tessler M, Cunningham SW, Twomey C, Amato G (2015) Integrating molecular, phenotypic and environmental data to elucidate patterns of crocodile hybridization in Belize. R Soc Open Sci 2:150409. https://doi.org/10.1098/rsos.150409
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Ahmed I (2011) Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071. https://doi.org/10.1093/bioinformatics/btr521
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Katoh K, Standley DM (2013) MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Res 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:356. https://doi.org/10.1186/s12859-014-0356-4
Lawson DJ, Hellenthal G, Myers S, Falush D (2012) Inference of population structure using dense haplotype data. PLoS Genet 8:e1002453. https://doi.org/10.1371/journal.pgen.1002453
Lee JC (2000) A field guide to amphibians and reptiles of the Maya World: the lowlands of México, Northern Guatemalan and Belize. Cornell University Press, New York
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. https://doi.org/10.1093/bioinformatics/btr509
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Malinsky M, Trucchi E, Lawson DJ, Falush D (2018) RADpainter and fineRADstructure: population inference from RADseq data. Mol Biol Evol 35:1284–1290. https://doi.org/10.1093/molbev/msy023
Mallet J (2005) Hybridization as an invasion of the genome. Trends Ecol Evol 20:229–237. https://doi.org/10.1016/j.tree.2005.02.010
Man Z, Yishu W, Peng Y, Xiaobing W (2011) Crocodilian phylogeny inferred from twelve mitochondrial protein-coding genes, with new complete mitochondrial genomic sequences for Crocodylus acutus and Crocodylus novaeguineae. Mol Phyl Evol 60:62–67. https://doi.org/10.1016/j.ympev.2011.03.029
Márdero S, Schmook B, Christman Z, Metcalfe SE, De la Barreda-Bautista B (2019) Recent disruptions in the timing and intensity of precipitation in Calakmul, Mexico. Theor Appl Climatol 140:129–144. https://doi.org/10.1007/s00704-019-03068-4
Meganathan P, Dubey B, Batzer MA, Ray DA, Haque I (2011) Complete mitochondrial genome sequences of three Crocodylus species and their comparison within the Order Crocodylia. Gene 478:35–41. https://doi.org/10.1016/j.gene.2011.01.012
Meredith RW, Hekkala ER, Amato G, Gatesy J (2011) A phylogenetic hypothesis for Crocodylus (Crocodylia) based on mitochondrial DNA: evidence for a trans-Atlantic voyage from Africa to the New World. Mol Phyl Evol 60:183–191. https://doi.org/10.1016/j.ympev.2011.03.026
Milian-Garcia Y, Jensen EL, Ribalta Mena S, Perez Fleitas E, Sosa Rodriguez G, Guerra Manchena L, Espinosa Lopez G, Russello MA (2016) Genetic evidence for multiple paternity in the critically endangered Cuban crocodile (Crocodylus rhombifer). Amph-Rept 37:273–281. https://doi.org/10.1163/15685381-00003056
Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA (2008) Database indexing for production MegaBLAST searches. Bioinformatics 24:1757–1764. https://doi.org/10.1093/bioinformatics/btn322
Muniz FL, Campos Z, Rangel SMH, Martínez JG, Souza BC, De Thoisy B, Botero-Arias R, Hrbek T, Farias IP (2018) Delimitation of evolutionary units in Cuvier’s dwarf caiman, Paleosuchus palpebrosus (Cuvier, 1807): insights from conservation of a broadly distributed species. Conserv Genet 19:599–610. https://doi.org/10.1007/s10592-017-1035-6
Muniz FL, Ximenes AM, Bittencourt PS, Hernández-Rangel SM, Campos Z, Hrbek T, Farias IP (2019) Detecting population structure of Paleosuchus trigonatus (Alligatoridae: Caimaninae) through microsatellites markers developed by next generation sequencing. Mol Biol Rep 46:2473–2484. https://doi.org/10.1007/s11033-019-04709-7
Nguyen TT, Ziegler T, Rauhaus A, Nguyen TQ, Tran DTA, Wayakone S, Luu VQ, Vences M, Le MD (2018) Genetic screening of siamese crocodiles (Crocodylus siamensis) in Laos and Vietnam: identifying purebred individuals for conservation and release programs. IUCN/SSC Crocodile Specialist Group Newsletter 37:8–14. http://www.iucncsg.org/365_docs/attachments/protarea/b0c9ec00c3b7e9d72ac6435d81f8320a.pdf.
Oaks JR (2011) A time-calibrated species tree of Crocodylia reveals a recent radiation of the true crocodiles. Evolution 65:3285–3297. https://doi.org/10.1111/j.1558-5646.2011.01373.x
Pacheco-Sierra G, Amavet PS (2021) Hybridization and speciation among new-world crocodilian species. In: Zucoloto RB, Amavet PS, Verdade LM, Farias IP (eds) Conservation genetics of new world crocodilians. Springer, Cham, pp 171–183
Pacheco-Sierra G, Gompert Z, Dominguez-Laso J, Vazquez-Dominguez E (2016) Genetic and morphological evidence of a geographically widespread hybrid zone between two crocodile species, Crocodylus acutus and Crocodylus moreletii. Mol Ecol 25:3484–3498. https://doi.org/10.1111/mec.13694
Pacheco-Sierra G, Vázquez-Domínguez E, Pérez-Alquicira J, Suárez-Atilano M, Domínguez-Laso J (2018) Ancestral hybridization yields evolutionary distinct hybrids lineages and species boundaries in crocodiles, posing unique conservation conundrums. Front Ecol Evol 6:138. https://doi.org/10.3389/fevo.2018.00138
Paris JR, Stevens JR, Catchen JM (2017) Lost in parameter space: a road map for stacks. Methods Ecol Evol 8:1360–1373. https://doi.org/10.1111/2041-210X.12775
Payseur BA, Rieseberg LH (2016) A genomic perspective on hybridization and speciation. Mol Ecol 25:2337–2360. https://doi.org/10.1111/mec.13557
Raj A, Stephens M, Pritchard JK (2014) fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197:573–589. https://doi.org/10.1534/genetics.114.164350
Ray DA, Dever JA, Platt SG, Rainwater TR, Finger AG, McMurry ST, Batzer MA, Barr B, Stafford PJ, McKnight J, Densmore LD III (2004) Low levels of nucleotide diversity in Crocodylus moreletii and evidence of hybridization with C. acutus. Conserv Genet 5:449–462. https://doi.org/10.1023/B:COGE.0000041024.96928.fe
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annu Rev Ecol Syst 27:83–109. https://doi.org/10.1146/annurev.ecolsys.27.1.83
Roberto IJ, Bittencourt PS, Muniz FL, Hernández-Rangel SM, Nóbrega YC, Ávila RW, Souza BC, Alvarez G, Miranda-Chumacero G, Campos Z, Farias IP, Hrbek T (2020) Unexpected but unsurprising lineage diversity within the most widespread Neotropical crocodilian genus Caiman (Crocodylia, Alligatoridae). Syst Biodiv 2:89. https://doi.org/10.1080/14772000.2020.1769222
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. https://doi.org/10.1038/nbt.1754
Rochette NC, Catchen JM (2017) Deriving genotypes from RAD-seq short-read data using Stacks. Nat Protocols 12:2640. https://doi.org/10.1038/nprot.2017.123
Rochette NC, Rivera-Colón AG, Catchen JM (2019) Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol Ecol 28:4737–4754. https://doi.org/10.1111/mec.15253
Rodriguez D, Cedeño-Vázquez JR, Forstner MRJ, Densmore LD III (2008) Hybridization between Crocodylus acutus and Crocodylus moreletii in the Yucatan Peninsula: II. Evidence from microsatellites. J Exp Zool A 309A:674–686. https://doi.org/10.1002/jez.499
Ross F, Mayer G (1983) On the dorsal armor of the Crocodilia. In: Rhodin AGJ, Miyata K (eds) Advances in herpetology and evolutionary biology. Museum of Comparative Zoology, Cambridge, pp 305–331
Ross CA, Ross FD (1974) Caudal scalation of central american Crocodylus. Proc Biol Soc Washington 87:231–234
RStudio Team (2020) RStudio: integrated development for R. pp. RStudio, PBC, Boston
Sánchez-Herrera O, Segurajáuregui GL, Ortiz de la Huerta AGN, Benítez Díaz H (2011) Programa de Monitoreo del Cocodrilo de Pantano (Crocodylus moreletii) México-Belice-Guatemala México. Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO), Mexico.
Serrano-Gomez SS, Guevara-Chumacero LM, Barriga-Sosa IDLA, Ulloa-Arvizu R, Gonzalez-Guzman S, Vazquez-Pelaez CG (2016) Low levels of genetic diversity in Crocodylus acutus in Oaxaca and Guerrero, Mexico, and molecular-morphological evidence of the presence of C. moreletii. Biochem Syst Ecol 69:51–59. https://doi.org/10.1016/j.bse.2016.08.005
Skotte L, Korneliussen TS, Albrechtsen A (2013) Estimating individual admixture proportions from next generation sequencing data. Genetics 195:693–702. https://doi.org/10.1534/genetics.113.154138
Soria-Ortiz GJ, García-Navarrete PG, Ochoa-Ochoa LM, Rincón-Gutiérrez A (2022) Potential distribution of hybrids between Crocodylus acutus and Crocodylus moreletii on the Mexican Pacific coast outside the natural hybridisation zone. Herpetol J 32:93–101. https://doi.org/10.33256/32.3.93101
Taylor SA, Larson EL (2019) Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat Ecol Evol 3:170–177. https://doi.org/10.1038/s41559-018-0777-y
Todesco M, Pascual MA, Owens GL, Ostevik KL, Moyers BT, Hübner S, Heredia SM, Hahn MA, Caseys C, Bock DG, Rieseberg LH (2016) Hybridization and extinction. Evol Appl 9:892–908. https://doi.org/10.1111/eva.12367
Toews DP, Brelsford A (2012) The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21:3907–3930. https://doi.org/10.1111/j.1365-294X.2012.05664.x
Truong HT, Ramos AM, Yalcin F, de Ruiter M, van der Poel HJA, Huvenaars KHJ, Hogers RCJ, van Enckevort LJG, Janssen A, van Orsouw NJ, van Eijk MJT (2012) Sequence-based genotyping for marker discovery and co-dominant scoring in germplasm and populations. PLoS ONE 7:e37565. https://doi.org/10.1371/journal.pone.0037565
Velo-Antón G, Godinho R, Campos JC, Brito JC (2014) Should I stay or should I go? Dispersal and population structure in small, isolated desert populations of West African crocodiles. PLoS ONE 9:e94626. https://doi.org/10.1371/journal.pone.0094626
Villela PMS, Coutinho LL, Piña CI, Verdade LM (2008) Macrogeographic genetic variation in broad-snouted caiman (Caiman latirostris). J Exp Zool A 309A:628–636. https://doi.org/10.1002/jez.489
vonHoldt BM, Brzeski KE, Wilcove DS, Rutledge LY (2018) Redefining the role of admixture and genomics in species conservation. Cons Lett 11:e12371. https://doi.org/10.1111/conl.12371
Winter DJ (2012) mmod: an R library for the calculation of population differentiation statistics. Mol Ecol Res 12:1158–1160. https://doi.org/10.1111/j.1755-0998.2012.03174.x
Wu TD, Reeder J, Lawrence M, Becker G, Brauer MJ (2016) GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. In: Mathé E, Davis S (eds) Statistical genomics: methods and protocols. Springer, New York, pp 283–334
Yamazaki D, Ikeshima D, Tawatari R, Yamaguchi T, O’Loughlin F, Neal JC, Sampson CC, Kanae S, Bates PD (2017) A high-accuracy map of global terrain elevations. Geophys Res Lett 44:5844–5853. https://doi.org/10.1002/2017GL072874
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. https://doi.org/10.1089/10665270050081478
Zucoloto RB, Farias IP, Amavet PS (2021) Molecular markers applied to conservation genetics of american crocodilians. In: Zucoloto RB, Amavet PS, Verdade LM, Farias IP (eds) Conservation genetics of new world crocodilians. Springer, Cham, pp 31–77
Acknowledgements
This study was possible thanks to the financial and logistical support of Operation Wallacea and the University of Salford through an ICase PhD sponsorship. Molecular bioinformatic analysis was carried at NERC’s Biomolecular Analysis Facility located in University of Sheffield thorough an NBAF support grant to JALBN and RJ. We would like to thank the communities of 16 de Septiembre, Álvaro Obrégon, Bel-Ha, Conhuas, Cristobal Colón, Dos Lagunas Norte, Dos Naciones, Flores Magón, Hormiguero, Keiche Las Pailas, Ley Fomento, Mancolona, Nuevo Becal and Valentín Goméz (Campeche, Mexico) for granting us the opportunity to work in their ejidal land and for providing guide services and logistical support. We thank O Platas-Vargas, L Hernandez, N Xe, SM Aguada, V Isoart, V Corradi, K Slater and C Acton for their assistance in the field. We also thank the IUCN-SSC Crocodile Specialist Group for granting the Student Research Assistance Scheme (SRAS) to JALBN; MA López-Luna and C Cedillo Leal for providing the external reference samples of C. moreletii and P Charruau for providing the external reference samples of C. acutus used in this study; A Lopez-Cen and D Sima-Pantí for their assistance with obtaining yearly research permits; SEMARNATCAM, J López-Sosa, A Balam-Koyoc, EL Hernández-Pérez, JG Hinterholzer Pino, SE Padilla and the team from Centro Operativo de las Direcciones de las Reservas Balam-Ku Balam-Kin for all their logistical support and assistance in fieldwork activities in Balam-Ku State Reserve; S Henderson, J Daw, JC Gutierrez, PE Nahuat-Cervera and the rest of the Operation Wallacea staff and student volunteers for their logistical support and assistance in the field.
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception and experimental design of this study. JALBN and RJ were involved in all steps of this research project and produced the first draft of this manuscript. Obtaining samples in the field was only possible due to the efforts of JALBN, MGJ, SPP and PC. Processing of samples in the laboratory, quality checks and material preparation for sequencing was carried by JALBN, AMA and JRCV. Bioinformatic analyses were carried out by JALBN, KM and RJ. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest nor have relevant financial or non-financial interests to disclose.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10592_2023_1544_MOESM1_ESM.jpeg
Supplementary file1 (JPEG 265 KB) Fig. S1 Six general sampling areas within Calakmul (white stars), and geographical origin of reference samples obtained of Crocodylus acutus (white triangles) and Crocodylus moreletii (red triangles) within Mexico (Pacheco Sierra et al., 2018, Pacheco Sierra et al., 2018).
10592_2023_1544_MOESM2_ESM.jpg
Supplementary file2 (JPG 2198 KB)Fig. S2 Visual assessment across our crocodile samples of the mapped sequences corresponding to mitochondrial genome regions (a) 2278_2872 and (b) 9528-9622 of Crocodylus moreletii (first row; NCBI Reference Sequence: NC_015235.1; Meganathan et al., 2011) and Crocodylus acutus (second row; NCBI Reference Sequence: NC_015647.1; Man et al., 2011) in TASSEL v.5.2 (Bradbury et al., 2007). Individuals highlighted within the red and black rectangles indicate, respectively, the external reference samples of C. acutus and C. moreletii used in this study.
10592_2023_1544_MOESM3_ESM.jpg
Supplementary file3 (JPG 302 KB) Fig. S3 Genetic structure and ancestry proportions (Q-scores) from NGSadmix (Skotte et al. 2013), with K = 2 to 6, where solid lines are 95% credible intervals and different colours represent each admixture proportion. Assignment results based on the CLUMPAK method (Kopelman et al. 2015) indicated that the number of clusters that best explained the structure was K = 4 (one cluster of C. acutus and three clusters of C. moreletii). Each vertical bar corresponds to one crocodile sample. Inverted black triangles represent samples with reads matching regions 2278-2872 and 9528-9622 of Crocodylus acutus mitochondrial genome. Dashed white lines delimit different sampled sites within each of the six general surveyed areas within the region of Calakmul (CK). CA and CM represent, respectively, the external reference samples of the C. acutus and C. moreletii.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Barão-Nóbrega, J.A.L., González-Jáuregui, M., Padilla-Paz, S. et al. Characterising a genetic stronghold amidst pervasive admixture: Morelet’s crocodiles (Crocodylus moreletii) in central Yucatan. Conserv Genet 24, 893–903 (2023). https://doi.org/10.1007/s10592-023-01544-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-023-01544-z