Abstract
The Kuranda Treefrog occurs in tropical north-east Australia and is listed as Critically Endangered due to its small distribution and population size, with observed declines due to drought and human-associated impacts to habitat. Field surveys identified marked population declines in the mid-2000s, culminating in very low abundance at most sites in 2005 and 2006, followed by limited recovery. Here, samples from before (2001–2004) and after (2007–2009) this decline were analysed using 7132 neutral genome-wide SNPs to assess genetic connectivity among breeding sites, genetic erosion, and effective population size. We found a high level of genetic connectivity among breeding sites, but also structuring between the population at the eastern end of the distribution (Jumrum Creek) versus all other sites. Despite finding no detectable sign of genetic erosion between the two times periods, we observed a marked decrease in effective population size (Ne), from 1720 individuals pre-decline to 818 post-decline. This mirrors the decline detected in the field census data, but the magnitude of the decline suggested by the genetic data is greater. We conclude that the current effective population size for the Kuranda Treefrog remains around 800 adults, split equally between Jumrum Creek and all other sites combined. The Jumrum Creek habitat requires formal protection. Connectivity among all other sites must be maintained and improved through continued replanting of rainforest, and it is imperative that impacts to stream flow and water quality are carefully managed to maintain or increase population sizes and prevent genetic erosion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endangered species are typically in population decline, with small and fragmented populations. This can drive a loss of genetic diversity and inbreeding, which in turn can generate further population decline, in what has been termed an extinction vortex (Gilpin & Soule 1986). The loss of genetic diversity, or ‘genetic erosion’, is a fundamental short- and long-term issue because it can reduce fitness and the capacity of populations to adapt to environmental change (Agashe et al. 2011; Berger et al. 2012). Inbreeding depression can become an added issue in small populations resulting in reduced survival and reproduction (Darwin 1876; O’Grady et al. 2006; Charlesworth and Willis 2009). Population fragmentation can further reduce genetic diversity within populations because effective population size is greatly reduced in each population, thus magnifying the potential effects of genetic drift and inbreeding depression (Frankham 1996; Aguilar et al. 2008). With limited gene flow, alleles lost in individual populations may not be reintroduced by migration, and may then be lost from the entire species when an individual population becomes small or goes extinct. Therefore, assessing and monitoring genetic erosion and connectivity in endangered species is necessary in order to conserve them.
Conservation genomics, using thousands of neutral genome-wide loci (typically, single-nucleotide polymorphisms, SNPs), greatly increased accuracy and fine-scale resolution for estimating parameters like population structuring, effective population size, demographic history, relatedness, and inbreeding (Luikart et al. 2003). To some degree, genomic approaches can also overcome the limitations of small sample sizes – typically associated with rare and endangered species – by providing deeper genetic insights per individual sampled (e.g., Shi et al. 2009; Willing et al. 2012). Identification of human-induced barriers to gene flow can guide conservation efforts, either via restoring habitat connectivity in the landscape or manually translocating individuals across barriers (Jha 2015; Ivy et al. 2016). Further, genetic data can be used to monitor genetic erosion and hence implement conservation actions promptly (Leroy et al. 2018). For instance, genome-wide estimates of effective population size through time can help detect population declines and genetic erosion early (Antao et al. 2011; O’Loughlin et al. 2016). Similarly, using metrics such as allelic richness can help detect the early stages of genetic erosion, when other metrics such as heterozygosity have not yet been affected (Hoban et al. 2014). Sampling populations at multiple points in time allows for detection of genetic erosion trends and the identification of possible underlying causes (Mathieu-Begne et al. 2019), especially when sampling is conducted pre- and post-disturbance (Vandergast et al. 2016).
Globally, amphibians are the most threatened vertebrate group, with approximately one-third of all described species on the IUCN red-list, including over 660 species listed as Critically Endangered and possibly more than 140 species extinctions since 1980 (Stuart et al. 2004; Scheele et al. 2017). Frog declines in Australia mirror the global trend, with six species listed as Extinct, 15 species listed as Critically Endangered, and 18 species listed as Endangered (IUCN 2021). One response to this has been detailed assessments of threats and potential management and research actions (e.g., Gillespie et al. 2020; Geyle et al. 2021), but these have been assessed at the whole species level and rarely considered genetic diversity and structuring within species. Conservation genomic studies have only been conducted on a handful of Australian frog species (Cummins et al. 2019; McKnight et al. 2019, 2020), and a detailed assessment of genetic diversity is lacking in most threatened Australian frog species. For example, conservation genomic studies have not been conducted for any of the 15 Critically Endangered (EPBC Act) Australian frog species. This is despite strong evidence that reversing genetic erosion via genetic management is one of the most effective ways to increase population size and adaptability of small and declining populations (Frankham 2015).
The Kuranda Treefrog (Litoria myola) is a stream-breeding rainforest frog restricted to a small area west of Cairns in north-east Australia (Fig. 1A). Males live and call from streamside vegetation near riffle zones, while females live away from the streams in the mid and upper strata of the forest. No data exists on the lifespan for this species, but based on aging of other Queensland rainforest frogs of broadly similar ecology (Morrison et al. 2004), most breeding adults are probably in the order of two to five years old. Females visit the streams to choose a male and lay approximately 500 eggs in the stream, which hatch to an aquatic tadpole stage (Hoskin 2007). Subadults are presumed to live in the mid and upper strata of the forest because they are rarely encountered along streams (Hoskin 2007). Litoria myola is listed as Critically Endangered on the IUCN Red List and on the Australian Federal (EPBC) and State (Queensland NCA) lists. These listings reflect the very small distribution (Extent of Occurrence 13.5 km2; Area of Occupancy 3.5 km2) and population size (estimates of 1000 adults or less), along with observed and projected declines due to drought and human-induced impacts on breeding sites (Hoskin 2007; DoE 2021). These impacts include forest clearing, impacts to streams such as damming, water extraction and sedimentation, and potentially invasive ants (Hoskin 2007, 2012; Hoskin et al. 2018; DoE 2021).
Surveys for calling males have found L. myola at 16 ‘breeding areas’, each defined as a separate stream (Fig. 1A; Hoskin 2007; Hoskin et al. 2018). Population declines were observed at all sites during a prolonged drought from 2003 to 2005 (Hoskin 2007). By 2005, L. myola had declined to low abundance at most sites and had effectively disappeared from one site where it had formerly been abundant (apart from an occasional calling male). The total census population estimate in the early 2000s, prior to declines, was approximately 1,000 adults (Hoskin 2007), but the estimate post-decline (i.e., after 2005) was around 700 adults (Hoskin 2007). Approximately half of the population was estimated to occur at Jumrum Creek (Fig. 1A) in both the pre- and post-decline estimates. While some populations gradually recovered after 2005, there have been more recent declines at some sites due to land-use impacts on rainforest and stream habitat (Hoskin et al. 2018; DoE 2021; Hoskin et al. unpub. data).
The current census estimate remains at about 700 adults, with approximately half at Jumrum Creek, moderate populations (100–200) estimated at two creeks (Warril Creek, Owen Creek; Fig. 1A), and populations at all other breeding sites estimated to be small (< 50) or very small (< 20) (Hoskin et al. 2018; Hoskin et al. unpub. data). Census population estimates are based on counts of adult males in sections of each breeding stream, extrapolated out to the known linear extent of the population on that stream, and then doubled to estimate the adult census population (i.e., assuming a 50:50 sex ratio). Importantly, the degree to which the species is fragmented into sub-populations is not known. Males have only been encountered along streams. Females are also typically found near streams but have occasionally been found in rainforest away from streams (Hoskin 2007). Given that breeding sites are discrete sections of different streams, and given that adults are primarily encountered at these sites, populations of the species have been assumed to be highly fragmented. Estimating connectivity between breeding sites has been identified as a key action for conservation of the species (Hoskin et al. 2018). Here we used a neutral genome-wide SNP dataset to:
- Aim 1:
-
Assess genetic population structure and connectivity between breeding sites. We predicted fine-scale structuring across the range of L. myola (at the individual stream level) due to the fact that the species is rarely observed in-between the discrete breeding sites.
- Aim 2:
-
Assess genetic diversity within each identified population. We expected moderate genetic diversity within the Jumrum Creek population due to the observed consistently ‘large’ population size over the last 20 years but lower genetic diversity within the other breeding sites due to small (and more variable) population sizes.
- Aim 3:
-
Estimate effective population size for the entire species and within genetically defined populations. Based on previous estimates of population size for the species, we predicted Jumrum Creek to have the largest effective population size, and all other populations to have smaller (< 200) effective population sizes.
- Aim 4:
-
Assess population size trends and genetic erosion by comparing estimates of effective population size and genetic diversity in two time periods of sampling. We expected to detect population declines and genetic erosion from the earlier to the later period based on observed population declines in the field.
Ultimately, we use our findings to fulfil key knowledge gaps identified for Litoria myola (Hoskin et al. 2018; DoE 2021), and outline on-ground conservation actions for this Critically Endangered species.
Methods
Sample Collection
A total of 126 Litoria myola individuals were sampled from 11 of the 16 known breeding streams, spanning the known distribution of L. myola at the time of sampling (Fig. 1A). Each breeding stream was sampled at one site, with the exception of three sites in the relatively large Jumrum Creek population and two sites on Owen Creek. Samples were obtained between 2001 and 2016, with two main sampling periods (2001–2004 and 2007–2009), followed by limited sampling in 2013 and 2016 (Table 2; Online resource 1). Extensive population declines had occurred by 2005, and some populations could not be sampled after the 2001–2004 period. We therefore used 2005 as the cut-off between pre- and post-decline. We conducted genetic analyses across three datasets: the time period (T) including all samples (T2001–2016), the main sampling period pre-decline (T2001–2004), and the main sampling period post-decline (T2007–2009).
Individuals were located while walking along streams, using either a head lamp to detect the frog’s eye reflection or by sound localization of male mating calls. A total of 105 males and 17 females were sampled. The two sexes are easily distinguishable by the presence of nuptial pads on males and their smaller body size. Tissue samples were collected by using sterile surgical scissors to remove a single toe pad from toe II (i.e., second innermost) on the right foot. Toe pads were placed in 95% ethanol. Litoria myola is morphologically similar to its sister species Litoria serrata, which co-occurs at nearly all sites. The majority of males can be identified in the field by call differences and smaller body size of L. myola but females are difficult to identify due to greater overlap in body size and their lack of vocalization (Hoskin 2007). The identification of all sampled individuals was thus checked using a diagnostic mtDNA marker (Hoskin et al. 2005).
Marker discovery, genotyping and filtering
Genomic DNA was extracted at Diversity Arrays Technology (DArT) Ltd. Canberra, Australia, using the Machery Nagel Tissue Kit with the Tecan 100 Robot, using a DArT proprietary extraction script. SNPs marker discovery and genotyping were conducted at the same facility with the DArTseq™ method (Sansaloni et al. 2011; Kilian et al. 2012; Lal et al. 2018). A digestion–ligation reaction was conducted at 37 °C for 2 h with ~ 100–200 ng of gDNA per individual, using a combination of Pstl and Sphl restriction enzymes and unique DArT proprietary barcodes. Barcodes were included in the Pstl-compatible adapter, alongside an Illumina flowcell attachment sequence and a sequencing primer. The reverse adapter contained a flowcell attachment region and an Sphl-compatible overhang sequence. For each sample, mixed fragments containing both Pstl and Sphl adapters were amplified through 30 rounds of PCR. Samples displaying incomplete or non-uniform digestion patterns when visualised through gel electrophoresis were discarded from the library. Equimolar amounts of PCR product for each sample were pooled into two batches and sequenced (77-bp single end reads) on a single flowcell per batch on an Illumina HiSeq2500, producing ~ 1.25 million raw reads per individual. Around 15% of individuals per batch were included twice in the libraries to produce random technical replicates. Genotypes were called with the DArT KDcompute pipeline (http://www.kddart.org/kdcompute.html), following Lind et al. 2017, as described for the closely related Green-eyed Treefrog (L. serrata) in McKnight et al. 2019. In brief, this involved demultiplexing using the unique barcodes, adapter trimming, removal of reads containing base-pair Q scores below 30, and removal of reads resulting from contamination by blasting raw reads to the DArTdb sequence database.
Upon receiving called genotype data from DArT, individuals with low Call Rate (i.e., sequenced for less than 80% of SNPs) or high heterozygosity (> 10%) were filtered because these could indicate low quality or contaminated samples. These thresholds were decided based on the distribution of individual call rate and heterozygosity, in order to remove obvious outliers. DArTseq™ SNPs were first filtered by call rate (i.e., the percentage of samples for which SNP information is not missing; CR > 90%) and monomorphic SNPs were removed manually in Microsoft Excel v16. Then, using the DARTQC filtering pipeline (https://github.com/esteinig/dartqc), genotype calls with less than 10 total reads were silenced, and SNPs were filtered by minor allele frequency (MAF, > 2%), CR (> 70%), and reproducibility (REP, > 90%). Here we used two call rate filtering steps to ensure a low error rate in the data. The first step, on the raw dataset, was the more stringent and the main call rate filtering step (CR > 90%). The second CR filtering step (CR > 70%) was used within DARTQC after silencing of individual calls to remove any marker with high proportions of missing data, which would not have been detected as a poor-quality marker ahead of silencing. Additionally, duplicates and clusters were filtered, retaining the SNP with the higher MAF. The DARTQC pipeline outputs data in the .ped format from PLINK, which was converted to alternative formats for downstream analyses using PGDSPIDER v.2.1.1.5 (Lischer and Excoffier 2011).
Full sibling pairs were identified using COANCESTRY v.1.0.1.10 (Wang 2011) and COLONY v.2.0.6.8 (Jones and Wang 2010), and from each pair only the sibling with the highest Call Rate was retained for Hardy-Weinberg Equilibrium (HWE) filtering. Before filtering for HWE and identifying outliers, a preliminary assessment of population structure was conducted with NETVIEW P (Neuditschko et al. 2012; Steinig et al. 2016; hereafter referred to as NETVIEW) and by discriminant analysis of principal components (DAPC) in the R package adegenet v.2.1.3 (Jombart et al. 2010; Jombart and Ahmed 2011), within R Studio v.1.3 (RStudio Team 2020), R v.4.0.3 (R Core Team 2021). Both methods identified two clusters, which were then used as populations for HWE filtering. The dataset was filtered for HWE equilibrium and linkage disequilibrium (LD), with an R2 value threshold of 0.6 using PLINK v.1.9 (Purcell et al. 2007). SNPs were then removed in an iterative fashion to retain one SNP per group of linked markers, as per McKnight et al. (2019).
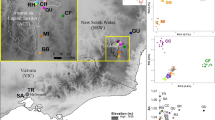
Population genetic structuring within subpopulations of the Kuranda Treefrog (Litoria myola). A Map showing the sampled range of the species, with occurrence records as white dots and sampling localities as coloured dots. Sampling localities abbreviations are provided in the legend. OF: Oak Forest; DC: Dismal Creek; QC: Queens Creek; NB: North Barron; CD: Cadagi Drive; OC: Owen Creek; WC: Warril Creek; NM: North Myola; BA: Barron Alambi; FA: Fairyland; JC: Jumrum Creek. NS represents sites where L. myola was detected but not sampled. The insets show the location of the study area within Queensland (QLD), Australia, and a photo of an adult male by Conrad Hoskin. The red line represents the Kennedy Highway. B, D, F. NETVIEW networks for the three time periods: (B) T2001–2016, (D) T2001–2004 and (F) T2007–2009. k-NN values are 25, 25 and 15 respectively. Individuals are coloured based on sampling locality. C, E, G. Structure plots for the three time periods: (C) T2001–2016, (E) T2001–2004 and (G) T2007–2009, for K values of 2, 2 and 3 respectively. Vertical black lines separate sampling localities, with the coloured dots underneath matching the localities in panel A
To identify outlier SNPs (i.e., markers that may be under selection), two different software were used: LOSITAN v.1.0 (Antao et al. 2008) and BAYESCAN v.2 (Foll 2012), which are based on different mathematical approaches and assumptions. LOSITAN was run for 50,000 generations with the following parameters: neutral mean FST, forcing mean FST, FDR of 0.1, and a confidence interval of 0.95. Five independent runs of LOSITAN were conducted, and SNPs were considered outliers if they were identified as such in all five runs. The GUI version of BAYESCAN was run with the following chain (sample size = 5,000; thinning interval = 10; pilot runs = 20; pilot runs length = 500; additional burn in = 50,000) and parameters (Prior odds for neutral model = 10; FIS uniform between 0 and 1). Any marker identified as an outlier by either method was removed from the neutral dataset. Finally, siblings previously removed were re-introduced in the dataset, and COANCESTRY and COLONY were used once more for sibling pair detection. From detected sibling pairs, only the sibling with the highest call rate was retained from each pair for downstream analyses, to avoid potential biases caused by the inclusion of closely related individuals (Devloo-Delva et al. 2019).
Genetic population structure and connectivity
From the overall filtered neutral dataset, three datasets were produced to assess genetic structuring and diversity indices for all samples (T2001–2016; n = 110) and to compare between the two intensively sampled periods: pre-decline (T2001–2004; n = 64) and post-decline (T2007–2009; n = 35). Analyses on the dataset with all samples (T2001–2016) were conducted to assess whether structuring patterns detected within the pre- and post-decline time periods were consistent and robust, given the small sample size and lack of temporal representations for some breeding sites in each time period (Online resource 1). Population genetic structure was then assessed for each of these datasets using three methods: population network analysis with NETVIEW (Neuditschko et al. 2012; Steinig et al. 2016); discriminant analysis of principal components (DAPC; Jombart et al. 2010); and the Bayesian clustering method of STRUCTURE v.2.3.4 (Pritchard et al. 2000). An analysis of molecular variance (AMOVA) and calculations of FST and PhiPT were implemented in GENALEX v.6.5.03 (Peakall and Smouse 2006). Details of each of these are below.
NETVIEW was used to build a k-nearest neighbours (k-NN) population network, after producing an identity by similarity distance matrix with PLINK for each of the three datasets. The k-NN network was constructed for a maximum number of nearest neighbours ranging between 5 and 25. DAPC was run on all three datasets, using the function find.clusters implemented in the adegenet package ( Jombart and Ahmed 2011). To identify the most likely cluster configuration in each of the three datasets, we used BIC as the measure of goodness of fit, and retained n/3 principal components, where n is the sample size for the dataset being used.
STRUCTURE was run on all three datasets, testing from 1 to 5 genetic clusters (K) with 3 replicates for each K (burn-in = 50,000; Markov chain Monte Carlo replicates = 300,000), with a fixed lambda value of 1, and inferring alpha. The ADMIXTURE model was adopted because the species originated recently in evolutionary time (Hoskin et al. 2005) and thus all individuals share recent common ancestors. Information about sampling locality was included with the LOCPRIOR option to account for the small sample size of some breeding sites and facilitate detection of clustering at the stream level if present. The ‘best’ value of K was identified based on both the L(K) and ΔK criteria (Pritchard et al. 2000; Evanno et al. 2005) using STRUCTURESELECTOR (Li and Liu 2018). To avoid detecting only the first major level of structuring, STRUCTURE was then run on the identified clusters independently with the same parameters as above. Finally, to ensure that detected clusters were not the result of uneven sample size, STRUCTURE was also run subsampling the larger Jumrum Creek population to 15 individuals, matching the sample size for the second and third largest sampled populations.
AMOVA statistics and differentiation indices were calculated in GENALEX for the three time periods, with 999 permutations and the samples divided into the most likely population clusters identified by DAPC, NETVIEW, and STRUCTURE.
Genetic diversity
Genetic diversity indices were estimated separately for each of the three time periods (i.e., T2001–2016, T2001–2004 and T2007–2009). For each time period, genetic diversity and genetic differentiation were estimated for the whole species, the identified genetic population clusters and the individual sites corresponding to distinct breeding streams. Average observed heterozygosity (Hobs), average expected heterozygosity corrected for sample size (Hn.b) and FIS were calculated in R, using the function gl.report.heterozygosity within the R package dartR v.2.0.4 (Gruber et al. 2018). Individual multi-locus heterozygosity (MLH) was estimated with the MLH function of the R package inbreedR v.0.3.2 and then averaged by population (Stoffel et al. 2016). Private alleles (PA) and allelic richness (AR) were estimated using HP-RARE v.1.0 (Kalinowski 2005), which applies a rarefaction method to account for uneven sample sizes. To allow comparisons both within and across time periods, rarefaction was implemented to match the smallest sample size at the finest hierarchical level (i.e., to match the population genetic cluster within a singular time period with the smallest sample size). Locus specific minor allele frequency was calculated using the minorAllele function of the adegenet package, and from this the proportion of rare alleles was obtained by dividing the number of SNPs with MAF < 0.05 by the total number of SNPs.
Effective population size
Effective population size (Ne) was estimated for all three time periods, to first estimate Ne for the entire species in the last twenty years, and then assess whether a change in effective population size could be detected between T2001–2004 and T2007–2009. Effective population size was calculated in NEESTIMATOR v.2.1 (Do et al. 2014) using the Linkage Disequilibrium (LD) estimator of Ne (NeLD) with random mating, using all standard critical values (i.e., 0.00, 0.01, 0.02, and 0.05) implemented in the software. We selected the LD method because it is able to detect even small population declines early when compared to the temporal method (Antao et al. 2011). Notably, this method estimates contemporary effective population size, which represents the Ne of the parent population, and not the sampled population. We ran five independent runs with random subsets of 1000 SNPs each to minimize the effect of physical linkage in our dataset. The number of SNPs was chosen as the lowest possible number still capable of achieving high precision of Ne estimates (Waples et al. 2016), to achieve a balance between minimizing linkage while retaining power. We then calculated the average of the five independent runs. Changing critical values had a minor effect on Ne estimates for the whole species and the Eastern cluster (Standard deviation < 100; Online resource 9), and therefore we report estimates for the more conservative critical value of 0.05. Additionally, we estimated Ne in two other ways, using the same software and parameters but changing the input data. Once using the full SNP dataset before LD filtering (n = 7,436) to assess whether filtering by LD hid the underlying biological signal, and once randomly subsampling populations for the T2001–2004 time period to match the sample size of time period T2007–2009, to assess whether any changes in Ne could be due to the difference in sample size between the two time periods.
Results
Marker discovery, genotyping and filtering
The initial raw dataset produced by DArT contained 60,815 SNPs across 126 Kuranda Treefrog (Litoria myola) individuals. Nine individuals were removed because of low Call Rate (CR) and one individual was removed because of high heterozygosity, leaving 116 samples. The first CR filter reduced the number to 31,822 SNPs. The dataset was further reduced to 9,890 SNPs following silencing by read depth and filtering by minor allele frequency, CR, and reproducibility, and removal of duplicates and clones. Filtering by HWE and LD left 7,215 SNPs (Online resource 2). Finally, BAYESCAN identified three outliers, while LOSITAN identified 83 outliers across all five independent runs, including those identified by BAYESCAN; resulting in a final dataset of 7,132 neutral SNPs. The same six full sibling pairs were identified by COLONY and COANCESTRY, both before and after filtering by HWE, LD, and removing outliers. All sibling pairs were detected within the same creek/sampling location, and were sampled within the same sampling event (Online resource 3). From each sibling pair, the individual with the highest call rate was retained for population structure analyses in STRUCTURE, DAPC and NETVIEW, for a total sample size of 110 individuals.
Genetic population structure and connectivity
STRUCTURE, DAPC, and NETVIEW analyses identified between one and three clusters across the three time periods investigated (Table 1), but two main geographically distinct clusters were consistently identified: an Eastern cluster formed by the samples from Jumrum Creek and half of the samples from the adjacent Fairyland (FA) site, and a Western cluster consisting of all remaining samples (Fig. 1). When analysing the two identified clusters separately, STRUCTURE did not identify any further clustering. The same two clusters were identified when subsampling the larger Jumrum Creek populations to account for uneven sampling. These results are discussed below, by time period, and a detailed summary containing STRUCTURESELECTOR plots, bar plots for each mode of K, BIC plots from the find.clusters function, and AMOVA results, is provided in the Supplementary Material (Online resource 4–8).
T2001–2016
When including all individuals sampled from 2001 to 2016 (n = 110), STRUCTURE identified K = 2 as the most likely value of K, both using the ΔK and LnP(K) criteria. DAPC also identified K = 2 as the most likely configuration. Both analyses split the sampled individuals into an Eastern cluster, comprising the Jumrum Creek samples (JC), and a Western cluster comprising remaining samples (Fig. 1B). The Fairyland (FA) population, for which only four individuals were sampled in total (all in 2016), is equally split between the two clusters. Two individuals sampled at JC2 are assigned to the Western cluster. The same pattern is observed in NETVIEW across all explored values of k-NN, and is most clear at k-NN = 25 (Fig. 1B). Pairwise FST and PhiPT between the two identified cluster at K = 2 were low, but significant, being 0.009 (p < 0.05) and 0.011 (p < 0.005), respectively. AMOVA also showed low among-cluster variance (1%).
T2001–2004
Samples collected between 2001 and 2004 show a similar pattern to the T2001–2016 dataset in terms of population structure. Analyses with STRUCTURE identified K = 2 as the best clustering using the ΔK method, and for the LnP(K) method K = 1 and K = 2 had nearly identical values with overlapping standard deviations. The same was true for DAPC, where K = 1 and K = 2 provided the best clustering and were equally likely. Once again, at K = 2 the JC population forms a separate (Eastern) cluster, while all remaining sampling localities cluster together (Fig. 1C). The split is discrete, with only one sample out of 30 from JC being assigned to the Western cluster. In NETVIEW plots, the Eastern and Western cluster are clearly visible but highly interconnected (Fig. 1C). As for the T2001–2016 analysis, pairwise Fst and PhiPT between the Eastern and Western cluster are low but significant (0.009, p < 0.05; 0.013, p < 0.01; respectively), and the variance explained among the two clusters is also low (1%).
T2007–2009
In the dataset comprising samples from 2007 to 2009, ΔK and LnP(K) found three and one clusters, respectively. The three clusters for the ΔK method represent the Western and Eastern cluster previously identified in the T2001–2016 and T2001–2004 time periods, as well as a third cluster consisting mainly of two individuals within the Eastern cluster (Fig. 1G). DAPC identified an optimal two clusters, representing the Eastern population comprising JC samples and a Western population comprising all remaining samples. NETVIEW clearly shows two clusters (the Eastern and Western) at k-NN = 15, but once again highly interconnected (Fig. 1F). FST and PhiPT values are higher than T2001–2004, but still low, at 0.011 (p < 0.05) and 0.019 (p < 0.01), respectively. AMOVA once again revealed that only 1% of variance is explained among the two clusters.
Genetic diversity
All genetic diversity measures investigated (Hobs, Hn.b., MLH, PA, AR, and the proportion of rare alleles) were very consistent both across populations and time periods (Table 2). Heterozygosity measures (Hobs, Hn.b., and MLH) were similar and often identical between the whole species and the identified Eastern and Western cluster, ranging between 0.30 and 0.32. Allelic richness (AR) ranged between 1.80 and 1.81, Private alleles (PA) were less than one for all populations after accounting for sample size, and the proportion of rare alleles (MAF < 0.05) ranged between 0.07 and 0.1 (Table 2). Heterozygosity metrics were consistent even for individual breeding streams, ranging between 0.28 and 0.30 (Hobs), and 0.29 and 0.32 (Hn.b.) (Online resource 14). Estimates of the proportion of rare alleles and FIS on the other hand were heavily affected by the small sample size at this scale and are thus not discussed here. All genetic diversity estimates are provided in Online resource 14.
Effective population size
Estimates for the whole species and the Eastern cluster had parametric 95% confidence intervals (CIs) below infinity (T2001–2016: Whole species CIs = 1,222–2,848; Eastern CIs = 681–4,109), except for one replicate (replicate A; Online resource 10). In contrast, Ne estimates for the Western cluster had wide CIs (T2001–2016: CIs = 1,544–29,321), and the upper CI extended to infinity in 12 of the 15 replicates runs (Online resource 10). Thus, we excluded any estimate with a parametric upper 95% CI reaching infinity, which indicates that results could be explained entirely by sampling error (Waples and Do 2010). All Ne estimates and 95% CIs are provided in the Supplementary Material and the averages from the five replicates are presented in Fig. 2.
Effective population size (Ne) and census population size (N) for the whole species and the Eastern genetic population cluster (essentially, Jumrum Creek). Ne estimates from this study are provided for all three time periods investigated (T2001–2016, T2001–2004 and T2007–2009). The average value from independent runs is reported, and individual estimates are displayed as black points. Point estimates of N were available only for time periods T2001–2004 and T2007–2009 (Hoskin 2007; Hoskin et al. 2018). Arrows highlight the detected decline between T2001–2004 and T2007–2009
Using all samples across the full time period (T2001–2016) gave an average Ne estimate for the whole species of 1,801; 1,335 for the Eastern cluster; and 2975 for the western cluster (Fig. 2). The estimates for T2001–2004 are similar for the whole species (1720), but lower for the Eastern and Western cluster (1009 and 1612, respectively). In contrast, the estimates for T2007–2009 are much lower for both the whole species (818) and the Eastern cluster (433), representing an over 50% decrease in both cases. For the Western cluster, two estimates reached infinity, and the remaining three had upper confidence limits reaching infinity. Therefore, for the Western cluster, a change in Ne between the two time periods could not be reliably assessed.
Waples and Do (2010) suggest using the lower CI as an indication of the lowest possible Ne bound for a population. This can be particularly useful when the estimates reach infinity. Lower CIs for estimates of Ne followed the same pattern of decline between T2001–2004 and T2007–2009, with a similar magnitude as that observed for the actual estimates of Ne (Online resource 10). Furthermore, using this approach we could assess possible changes in Ne in the Western cluster, for which no reliable Ne estimates could be obtained for T2007–2009. This revealed once again a decline in Ne between the two time periods, although with lower magnitude (~ 22% decline; Online resource 10). Estimating Ne with both the full SNP dataset before filtering for LD, and subsampling individuals for T2001–2004 to match the sample size for T2007–2009, did not change the observed pattern of decline (Online resources 10–11).
Discussion
We analysed a large genomic dataset of 7132 neutral SNPs for 110 individuals to guide conservation actions for the Critically Endangered Kuranda Treefrog (Litoria myola). Based on field observations, we expected individual breeding populations to be highly isolated, declining and experiencing genetic erosion. While we detected only moderate genetic population structure and good connectivity across many breeding populations, we also detected sharp and rapid population declines. These declines had not resulted in genetic erosion at the time of sampling. Below, we further discuss these results, and highlight their implication for the conservation of the species.
Genetic population structure and connectivity
Our first aim was to determine connectivity between known breeding sites. Because the species is rarely observed away from discrete sections of streams, we predicted fine-scale structuring at the individual stream level across the range of L. myola. We did find genetic structuring at the stream level, but only of Jumrum Creek in the east of the range versus the remaining creeks to the west (which were themselves genetically indistinguishable from each other in all analyses). The two genetic clusters—Eastern (Jumrum Creek) and Western—were recovered in all analyses. The Western cluster included all samples from the nine streams sampled across the western 80% of the range. The geographically intermediate Fairyland site (FA; yellow symbol) was found to be evenly genetically mixed between the Eastern and Western clusters (Fig. 1), and hence falls in the area of connectivity between the two genetic groups. These results suggest reduced gene flow between Jumrum Creek and all other sites, with Fairyland representing an intermediate population.
In spite of the consistent recovery of an Eastern and Western cluster, genetic differentiation between these two clusters was low (FST ~ 0.01) and we detected movement of two individuals between the clusters (discussed below). An important question is whether the Eastern versus Western genetic structuring reflects restricted gene flow due to natural structuring (i.e., as a result of natural processes over hundreds or thousands of years), or human-induced impediments acting either currently or historically (i.e., within the last 140 years of intense human use of the region). In considering structuring due to natural causes, the genetic structuring may reflect that Jumrum Creek is at the far eastern end of the range; however, in comparison, the site equally geographically separated at the far west of the range (Oak Forest, OF) is not genetically distinct (but note that we only had two samples from that site). Prior to modern settlement, the Kuranda area was fairly continuous rainforest, particularly along the Barron River and associated creeks, and the area between the Eastern and Western clusters would have been no exception. A study on the closely related L. serrata, which is 1% divergent for mtDNA from L. myola (Hoskin et al. 2005), found no fine-scale structuring in areas of fairly continuous rainforest for well surveyed populations that have retained constant population size since the 1980s (McKnight et al. 2019). Additionally, there is no sign that the Barron River has been a barrier to dispersal of L. myola, with no structuring detected between breeding sites on the northern versus southern side (Fig. 1).
We therefore suggest that the genetic structuring within Litoria myola reflects human-induced impacts on genetic connectivity in the last 140 years, with land clearing as the primary cause. The Kuranda region was settled by European settlers in the 1880s, and has had a history of logging, dairy and beef cattle farming, and coffee plantations. Today the land use is residential within the Kuranda township, and primarily rural residential outside of this (Fig. 1). Much of the current rainforest habitat in the Kuranda region is regrowth, with only small areas and riparian strips of remnant (uncleared) habitat. Assessment of aerial imagery shows that there is substantially more rainforest in some parts of the distribution of Litoria myola currently than there was in the first half of the 20th century. For example, aerial imagery from the year 1952 shows that the area between Jumrum Creek (Eastern) and the Western genetic cluster was heavily cleared (i.e., Kuranda township, Fairyland, and Myola areas) (Online resource 12). This clearing extended right to the southern bank of the Barron River, particularly between Warril Creek (WC) and Jumrum Creek, which potentially severed natural genetic connectivity for decades. The Eastern versus Western genetic structuring is consistent with this historic land clearing. Although roads have been shown to reduce gene flow and generate structuring in amphibian populations only 2 km apart (e.g., tiger salamander, McCartney-Melstad et al. 2018), across similar geographic scales to our study, we suggest the impact on gene flow is a result of historic land clearing rather than reduced connectivity due to roads, because the only major road through the L. myola distribution (Fig. 1A) is raised above tree height where it goes through the habitat, and is thus unlikely to represent a barrier to the frogs.
Apart from the presence of Eastern and Western clusters, our results show much higher connectivity than we predicted. Samples from streams across the entire western 80% of the range are genetically indistinguishable, even using these high-resolution markers. Further, we detected probable evidence of current movement between the two clusters, with two individuals sampled at Jumrum Creek clustering among the Western samples (Fig. 1B). This male and female were of Western genetic origin but were sampled at Jumrum Creek, suggesting a journey of at least 3 km. Adult females are regularly encountered in the forest and have been found up to 100 m from streams, which is towards the maximum distance that surveys have been conducted from streams (Hoskin 2007; Hoskin et al. 2018). It is therefore not surprising that they could travel between some streams. In contrast, males have only ever been encountered adjacent to streams, so it is surprising that one of the two long distance movements that we detected was a male.
Interestingly, we detected six full sibling pairs, with each sibling pair being adult frogs from the same sampling location and sampling event. As would be expected, the sibling pairs were detected at the four sites that had the largest sampling effort (JC1, JC2 WC, QC) (Online Resources 1, 3). However, the number of full sibling pairs is striking. For example, the eleven individuals sampled at site JC1 in 2001 included three full sibling pairs. Five of the six sibling pairs found across all sites involved males, with only one involving a male and a female. Based on rainforest species of similar broad ecology and size (Morrison et al. 2004), Litoria myola probably reaches breeding age at two or three years old, so the detected sibling pairs in our sample were presumably at least this age. The number of full sibling adults in our sample, with the frogs in each pair found in close proximity, suggests that Litoria myola has generally low dispersal (at least for males).
Further research is required to estimate dispersal distances, and whether movement between breeding sites is typically by females, males, and/or sub-adult frogs. Nevertheless, the results so far suggest that the species may generally not disperse far, but that some long-distance movements are possible and sufficient to retain connectivity across breeding streams. Importantly, this movement is unlikely to occur through non-forested habitat. Despite natural regrowth and active replanting of rainforest in recent decades, extensive cleared areas remain between most streams. The riparian habitat corridor along the Barron River probably acts as the primary dispersal pathway for adult frogs. However, the river itself is not likely to be a movement pathway. This is because the adult frogs are not aquatic, and the aquatic eggs and tadpoles would have limited survival in the river due to the abundance of predatory fish. Further, the Barron River flows west to east, so if egg or tadpole dispersal along the Barron River was prevalent, Jumrum Creek would be unlikely to be genetically distinct, being located at the far eastern (i.e., downstream) end of the distribution.
Genetic diversity
Our second aim was to assess genetic diversity across the whole species and any genetically identified population. Our prediction was that the Jumrum Creek population would have at least moderate genetic diversity due to the relatively large population size (in the order of hundreds of adults) observed over the years in field surveys, whereas the other breeding sites would have low genetic diversity due to small and, in some cases, declining population sizes. Unexpectedly, we found moderate heterozygosity (i.e., Hobs, Hn.b., and MLH ≈ 0.3) for the two genetic clusters identified, the whole species (Table 2) and the individual breeding streams (Online resource 14). We also found no observable change in heterozygosity, allelic richness and number of private alleles between the two well-sampled time periods T2001–2004 and T2007–2009 (Table 2). The observed heterozygosity in L. myola (and populations within) is comparable to that observed (also using DArTseq™ SNP data) in the relatively widespread and abundant sister species, L. serrata (McKnight et al. 2019), and double the heterozygosity in another Australian hylid (Litoria ewingii, Melville et al. 2017) and an Australian myobatrachid frog (Pseudophryne guentheri, Cummins et al. 2019). High genetic diversity at smaller breeding sites of L. myola is surprising, but can be explained by the high level of connectivity detected between them. Rather than acting as many small, independent populations, these are all likely to be connected as a relatively large Western population, and therefore able to retain genetic diversity. It is important to note that sample size for many of the individual breeding sites sampled post-drought was small (N < = 5; Online resource 1). Therefore, it is possible that while the Western cluster as a whole retained good levels of genetic diversity, some breeding sites within this cluster may have experienced genetic diversity loss that could not be detected with our sample sizes.
Effective population size
Our third aim was to estimate effective population size (Ne) of the whole species and the identified populations. We could not obtain consistent estimates for the Western cluster for T2007–2009, so here we focus on estimates for the whole species and the Eastern cluster (for whom estimates of Ne were consistent across replicates and only once had an upper CI reaching infinity). For T2001–2016, average Ne was 1,801 and 1,335 for the whole species and Eastern cluster, respectively. In T2001–2004, Ne estimates were similar, at 1,720 and 1,009 individuals for the wholes species and Eastern cluster, respectively. A marked decline in Ne was observed for the later time period T2007–2009, for which Ne estimates dropped to 818 for the whole species and 434 for the Eastern cluster, representing a 55% and 68% decline respectively (discussed below). Effective population size estimates were larger than our field census population size estimates, particularly for the pre-decline period (T2001–2004) (Fig. 2). While this result used to be uncommon, with most early studies finding Ne/N ratios of < 0.5 (reviewed in Frankham 1995; Palstra and Ruzzante 2008), more recent work has found many examples with Ne/N ratios above 1, and up to 1.7 in some amphibians (Waples et al. 2013). See Online resource 13 for a discussion of the potential reasons for the high Ne/N ratio in this study. The current population size for Litoria myola probably remains similar to the T2007–2009 Ne estimates; i.e., less than 800 individuals, with about half of these at Jumrum Creek. Field surveys conducted after the time period investigated here showed that some populations have recovered slightly after 2010 (but none to pre-decline densities), while the two largest populations have declined even further (Jumrum Creek, JC; Warril Creek, WC. Hoskin et al. unpub. data).
The reason for Ne estimates for the Western cluster reaching infinity remains unclear. Four main sources of bias are suggested for the linkage disequilibrium estimator of Ne that can result in inflated estimates: (1) difficulty in achieving high precision when the true Ne is larger than 1000 (Waples and Do 2010); (2) rare alleles (Turner et al. 2001; Waples and Do 2010); (3) allelic dropout (Russell and Fewster 2009): and (4) high migration between relatively small populations within a metapopulation. The first source of bias is well documented (e.g., Waples and Do 2010), but is unlikely to be the cause as it does not explain why Ne estimates were consistent and had narrow CIs in the whole species and the Eastern cluster and not the Western cluster (all of which had Ne estimates > 1000); nor does it explain why the estimates for the Western cluster are larger than that of the whole species (which includes all the Western cluster individuals within the estimate). The second source of bias, inflation due to rare alleles, is usually addressed by adjusting the value of Pcrit. Waples and Do (2010) in particular recommend that for sample sizes (S) below 25, a Pcrit value larger than 1/(2 S) and smaller than 1/S should be used. For the T2007–2009 Western cluster estimate, the sample size is 17, thus our adopted Pcrit value of 0.05 falls within the recommended range [1/(2*17) = 0.03, and 1/17 = 0.06]. The third possible source of bias, allelic dropout, might also have upwardly biased our Ne estimates; indeed data from mating crosses obtained for this species, using the same methodology as the one adopted here, did reveal medium rates of allelic dropout (Bertola et al. unpub. data). However, once again, that would be a shared issue across all the estimates, rather than for the Western cluster alone.
The fourth possible source of bias is a high level of migration between small populations within a metapopulation. In this scenario, small sample sizes for individual subpopulations can inflate Ne estimates (Waples and Do 2010). If we consider the Western cluster a metapopulation, with individual breeding sites acting as small populations but experiencing high levels of gene flow between each other, it could explain the inflated Ne estimates obtained. If this was the case, we would expect the issue of inflated Ne estimates to be more pronounced for T2007–2009, for which sample sizes of individual breeding sites are the smallest (n = 2–8). This is indeed the case, with all five Ne estimates for the Western cluster for T2007–2009 having an upper CI at infinity, and two estimates reaching infinity themselves (Online resource 10). On the other hand, results presented here point to the Western cluster being a highly interconnected population, with individual streams being genetically indistinguishable from each other. In light of this, one could argue about whether the Western cluster represents a metapopulation with high gene flow or a single homogeneous population. If the latter is true, then the sample size for this population is comparable to the Eastern cluster, and this possible source of bias does not explain the observed inflated Ne estimates. Hence, the reason behind the inflated Ne estimates for the Western cluster remains unclear. Therefore, we focus on estimates for the whole species and the Eastern cluster.
Population decline and genetic erosion
Our final aim was to assess population decline and genetic erosion before and after 2005. Estimates of Ne detected a substantial decline, mirroring the decline detected by census population size estimates (Hoskin 2007; Hoskin et al. 2018). While there are potential biases in both Ne and census estimates, the trend of decline is similar in both, and shared for the whole species and the Eastern cluster (Fig. 2). While we could not assess a decline in Ne estimates for the Western cluster directly, an assessment of lower CIs still supported a decline in Ne, with an estimated reduction of 22% (Online resource 10). In stark contrast to the estimated declines in Ne, heterozygosity and allelic richness estimates did not decline from T2001–2004 to T2007–2009 for the species as a whole or for either genetic cluster (Table 2). Given that contemporary Ne estimated here represents the Ne of the parent population (rather than the sampled population), it is even more surprising that no decline in other genetic diversity metrics could be detected in the sampled population.
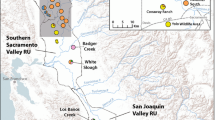
Visual summary of management recommendations for Litoria myola. All known records for the species are depicted as circles, with coloured circles representing breeding sites sampled for this study. Colours of sampled breeding sites match Fig. 1A. Green symbols are Jumrum Creek, yellow is the Fairyland site, and the remaining coloured sites form the Western genetic cluster. The red line shows the Kennedy Highway and blue lines represent streams within the region. The thicker blue line is the Barron River, the main waterbody in the region
Overall, our results suggest recent, rapid and sharp population declines in Litoria myola, without genetic erosion at the time of sampling. This concurs with studies suggesting that while changes in effective population size can be detected as early as ~ 5 generations after the decline (Antao et al. 2011; Hollenbeck et al. 2016), immediate genetic diversity loss may not occur unless the final population size is below ~ 100 individuals (Hoban et al. 2014). These results add to other studies showing that, with sufficient temporal sampling, genomic data can aid to detect population declines in its early stages (e.g., Antao et al. 2011; Skrbinšek et al. 2012; Hollenbeck et al. 2016; Vandergast et al. 2016). Importantly, more than ten years have passed since the last thorough sampling of this species (T2007–2009), and field surveys have detected continued declines. Thus, it is possible that genetic erosion has taken place since, warranting prompt conservation actions (discussed below).
Management recommendations
From our genetic results, we suggest the following on-ground conservation actions (Fig. 3):
-
(1)
Jumrum Creek harbours approximately half the population of Litoria myola and contains unique genetic diversity; hence, all efforts must be made to maintain the large population at this site. This includes formally protecting the main area of breeding habitat, which is mostly in unprotected land downstream of Jumrum Creek Conservation Park; and managing impacts to water flow and water quality from activities in the large and increasingly urbanized catchment.
-
(2)
Populations on all the other streams form the Western genetic cluster (with the exception of FA as a mixed site) and all of these likely play a role as stepping stones in maintaining connectivity across this cluster. However, a number of these populations are currently extremely small and under threat from ground water extraction, sedimentation, clearing, urban development, stream damming and, potentially, invasive ants (Hoskin 2007, 2012; Hoskin et al. 2018; DoE 2021). Threats to each breeding site need to be identified and minimised in order to maintain and improve each of these populations.
-
(3)
Our results support efforts to improve rainforest connectivity along the Barron River vegetation corridor. In particular, our results suggest replanting efforts would be well spent between the Western and Eastern clusters (i.e., between Warril Creek, Fairyland, and Jumrum Creek). This corridor exists due to replanting efforts by the community group Kuranda EnviroCare and private landowners, and these efforts should be supported into the future. Much of this land is owned by the Queensland Department of Transport and Main Roads and the Mareeba Shire Council, meaning these groups are key stakeholders in the future of this Critically Endangered species.
-
(4)
Further surveys should be conducted across the range of the species and in nearby streams in order to better resolve the distribution of Litoria myola. This includes thorough surveys of Flaggy Creek to estimate the extent and size of the recently discovered population there. Genetic analysis of this population would be valuable to assess whether it is part of the Western genetic cluster or is genetically distinct.
-
(5)
Continued field monitoring of key populations to assess population trends and detect further declines (or recovery) at breeding sites. Key sites include Jumrum Creek, Warril Creek, Owen Creek, and several of the very small populations in the Western cluster. Should significant declines be detected, translocation of individuals between the Eastern and Western cluster should be considered in an attempt to off-set loss of genetic diversity within each.
Data availability
The demultiplexed raw sequence data is available on the NCBI sequence read archive (BioProject PRJNA868494, Accessions SRR21074468–SRR21074622) and the filtered neutral dataset containing 7,132 SNPs and 110 individuals is available through FigShare (https://doi.org/10.6084/m9.figshare.20493423).
Code availability
Not applicable.
Change history
24 February 2023
Missing Open Access funding information has been added in the Funding Note.
References
Agashe D, Falk JJ, Bolnick DI (2011) Effects of founding genetic variation on adaptation to a novel resource. Evolution 65:2481–2491. https://doi.org/10.1111/j.1558-5646.2011.01307.x
Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Lobo J (2008) Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol Ecol 17:5177–5188. https://doi.org/10.1111/j.1365-294X.2008.03971.x
Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G (2008) LOSITAN: a workbench to detect molecular adaptation based on a F ST-outlier method. BMC Bioinformatics 9:323. https://doi.org/10.1186/1471-2105-9-323
Antao T, Pérez-Figueroa A, Luikart G (2011) Early detection of population declines: high power of genetic monitoring using effective population size estimators. Evol Appl 4:144–154. https://doi.org/10.1111/j.1752-4571.2010.00150.x
Berger JD, Buirchell BJ, Luckett DJ, Nelson MN (2012) Domestication bottlenecks limit genetic diversity and constrain adaptation in narrow-leafed lupin (Lupinus angustifolius L.). Theor Appl Genet 124:637–652. https://doi.org/10.1007/s00122-011-1736-z
Charlesworth D, Willis JH (2009) The genetics of inbreeding depression. Nat Rev Genet 10:783. https://doi.org/10.1038/nrg2664
Cummins D, Kennington WJ, Rudin-Bitterli T, Mitchell NJ (2019) A genome‐wide search for local adaptation in a terrestrial‐breeding frog reveals vulnerability to climate change. Glob Change Biol 25:3151–3162. https://doi.org/10.1111/gcb.14703
Darwin C (1876) The effects of cross and self fertilisation in the vegetable kingdom. Appleton and Company, New York
Department of the Environment (2021) Litoria myola in Species Profile and Threats Database, Department of the Environment, Canberra. Available from: https://www.environment.gov.au/sprat. Accessed Tue, 9 Nov 2021 18:42:26 + 1100
Devloo-Delva F, Maes GE, Hernández SI, Mcallister JD, Gunasekera RM, Grewe PM, Thomson RB, Feutry P (2019) Accounting for kin sampling reveals genetic connectivity in Tasmanian and New Zealand school sharks, Galeorhinus galeus. Ecol Evol 9:4465–4472. https://doi.org/10.1002/ece3.5012
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (ne) from genetic data. Mol Ecol Resour 14:209–214. https://doi.org/10.1111/1755-0998.12157
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Foll M (2012) BayeScan v2.1 user manual. Ecology 20:1450–1462
Frankham R (1995) Effective population size/adult population size ratios in wildlife: a review. Genet Res 66:95–107. https://doi.org/10.1017/S0016672300034455
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508. https://doi.org/10.1046/j.1523-1739.1996.10061500.x
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618. https://doi.org/10.1111/mec.13139
Geyle HM, Hoskin CJ, Bower DS et al (2021) Red hot frogs: identifying the australian frogs most at risk of extinction. Pac Conserv Biol. https://doi.org/10.1071/PC21019
Gillespie GR, Roberts JD, Hunter D, Hoskin CJ, Alford RA, Heard GW, Hines H, Lemckert F, Newell D, Scheele BC (2020) Status and priority conservation actions for australian frog species. Biol Conserv 247:108543. https://doi.org/10.1016/j.biocon.2020.108543
Gilpin ME, Soulé ME (1986) Minimum viable populations: processes of extinction. In: Soulé ME (ed) Conservation Biology: the Science of Scarcity and Diversity. Sinauer Associates, Sunderland, MA, pp 19–34
Gruber B, Unmack PJ, Berry OF, Georges A (2018) dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol Ecol Resour 18:691–699. https://doi.org/10.1111/1755-0998.12745
Hoban S, Arntzen JA, Bruford MW, Godoy JA, Rus Hoelzel A, Segelbacher G, Vilà C, Bertorelle G (2014) Comparative evaluation of potential indicators and temporal sampling protocols for monitoring genetic erosion. Evol Appl 7:984–998. https://doi.org/10.1111/eva.12197
Hollenbeck CM, Portnoy DS, Gold JR (2016) A method for detecting recent changes in contemporary effective population size from linkage disequilibrium at linked and unlinked loci. Heredity 117:07–216. https://doi.org/10.1038/hdy.2016.30
Hoskin CJ (2007) Description, biology and conservation of a new species of australian tree frog (Amphibia: Anura: Hylidae: Litoria) and an assessment of the remaining populations of Litoria genimaculata Horst, 1883: systematic and conservation implications of an unusual speciation event. Biol J Linn Soc 91:549–563. https://doi.org/10.1111/j.1095-8312.2007.00805.x
Hoskin CJ (2012) Kuranda Treefrog (Litoria myola). In: Curtis LK, Dennis AJ, McDonald KR, Kyne PM, Debus SJS (eds) Queensland’s threatened species. CSIRO Publishing, pp 156–157
Hoskin CJ, Higgie M, McDonald KR, Moritz C (2005) Reinforcement drives rapid allopatric speciation. Nature 437:1353–1356. https://doi.org/10.1038/nature04004
Hoskin CJ, Sharry P, Wannan B, Hepburn L, Shee R, Zehntner M, Donald D, Retter C, Smith K (2018) Community Action Plan for the conservation of the Kuranda Tree Frog (Litoria myola) and its Habitat 2018–2023. Kuranda, Australia. http://www.envirocare.org.au/frog-habitat-project.html
IUCN (2021) The IUCN Red List of Threatened Species. Version 2021-2. https://www.iucnredlist.org. Downloaded on 09 of November, 2021
Ivy JA, Putnam AS, Navarro AY, Gurr J, Ryder OA (2016) Applying SNP-derived molecular coancestry estimates to captive breeding programs. J Hered 107:403–412. https://doi.org/10.1093/jhered/esw029
Jha S (2015) Contemporary human-altered landscapes and oceanic barriers reduce bumble bee gene flow. Mol Ecol 24:993–1006. https://doi.org/10.1111/mec.13090
Jombart T, Ahmed I (2011) Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071. https://doi.org/10.1093/bioinformatics/btr521
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Jones OR, Wang J (2010) COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 10:551–555. https://doi.org/10.1111/j.1755-0998.2009.02787.x
Kalinowski ST (2005) hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska K, Jaccoud D, Hopper C, Aschenbrenner-Kilian M, Evers M, Peng K, Cayla C, Hok P, Uszynski G (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. Data production and analysis in population genomics. Humana Press, Totowa, NJ, pp 67–89. https://doi.org/10.1007/978-1-61779-870-2_5
Lal MM, Southgate PC, Jerry DR, Zenger KR (2018) Genome-wide comparisons reveal evidence for a species complex in the black-lip pearl oyster Pinctada margaritifera (Bivalvia: Pteriidae). Sci Rep-UK 8:191. https://doi.org/10.1038/s41598-017-18602-5
Leroy G, Carroll EL, Bruford MW, DeWoody JA, Strand A, Waits L, Wang J (2018) Next-generation metrics for monitoring genetic erosion within populations of conservation concern. Evol Appl 11:1066–1083. https://doi.org/10.1111/eva.12564
Lind CE, Kilian A, Benzie JAH (2017) Development of diversity arrays technology markers as a tool for rapid genomic assessment in Nile tilapia, Oreochromis niloticus. Anim Genet 48:362–364. https://doi.org/10.1111/age.12536
Lischer HE, Excoffier L (2011) PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28:298–299. https://doi.org/10.1093/bioinformatics/btr642
Li YL, Liu JX (2018) StructureSelector: a web-based software to select and visualize the optimal number of clusters using multiple methods. Mol Ecol Resour 18:176–177. https://doi.org/10.1111/1755-0998.12719
Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4:981. https://doi.org/10.1038/nrg1226
Mathieu-Bégné E, Loot G, Chevalier M, Paz‐Vinas I, Blanchet S (2019) Demographic and genetic collapses in spatially structured populations: insights from a long‐term survey in wild fish metapopulations. Oikos 128:196–207. https://doi.org/10.1111/oik.05511
McCartney-Melstad E, Vu JK, Shaffer HB (2018) Genomic data recover previously undetectable fragmentation effects in an endangered amphibian. Mol Ecol 27:4430–4443. https://doi.org/10.1111/mec.14892
McKnight DT, Lal MM, Bower DS, Schwarzkopf L, Alford RA, Zenger KR (2019) The return of the frogs: the importance of habitat refugia in maintaining diversity during a disease outbreak. Mol Ecol 28:2731–2745. https://doi.org/10.1111/mec.15108
McKnight DT, Carr LJ, Bower DS, Schwarzkopf L, Alford RA, Zenger KR (2020) Infection dynamics, dispersal, and adaptation: understanding the lack of recovery in a remnant frog population following a disease outbreak. Heredity 125:110–123. https://doi.org/10.1038/s41437-020-0324-x
Melville J, Haines ML, Boysen K, Hodkinson L, Kilian A, Smith Date KL, Potvin DA, Parris KM (2017) Identifying hybridization and admixture using SNPs: application of the DArTseq platform in phylogeographic research on vertebrates. Royal Soc Open Sci 4:161061. https://doi.org/10.1098/rsos.161061
Morrison C, Hero JM, Browning J (2004) Altitudinal variation in the age at maturity, longevity, and reproductive lifespan of anurans in subtropical Queensland. Herpetologica 60:34–44. https://doi.org/10.1655/02-68
Neuditschko M, Khatkar MS, Raadsma HW (2012) NETVIEW: a high-definition network-visualization approach to detect fine-scale population structures from genome-wide patterns of variation. PLoS ONE 7:e48375. https://doi.org/10.1371/journal.pone.0048375
O’Grady JJ, Brook BW, Reed DH, Ballou JD, Tonkyn DW, Frankham R (2006) Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol Conserv 133:42–51. https://doi.org/10.1016/j.biocon.2006.05.016
O’Loughlin SM, Magesa SM, Mbogo C, Mosha F, Midega J, Burt A (2016) Genomic signatures of population decline in the malaria mosquito Anopheles gambiae. Malar J 15:182. https://doi.org/10.1186/s12936-016-1214-9
Palstra FP, Ruzzante DE (2008) Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol Ecol 17:3428–3447. https://doi.org/10.1111/j.1365-294X.2008.03842.x
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
RStudio Team, RStudio (2020) RStudio: Integrated Development for R. PBC, Boston, MA URL. http://www.rstudio.com/
Russell JC, Fewster RM (2009) Evaluation of the linkage disequilibrium method for estimating effective population size. Modeling demographic processes in marked populations. Springer, Boston, pp 291–320. https://doi.org/10.1007/978-0-387-78151-8_13
Sansaloni C, Petroli C, Jaccoud D, Carling J, Detering F, Grattapaglia D, Kilian A (2011) Diversity arrays technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. https://doi.org/10.1186/1753-6561-5-S7-P54
Scheele BC, Skerratt LF, Grogan LF, Hunter DA, Clemann N, McFadden M, Newell D, Hoskin CJ, Gillespie GR, Heard GW, Brannelly L, Roberts AA, Berger L (2017) After the epidemic: ongoing declines, stabilizations and recoveries in amphibians afflicted by chytridiomycosis. Biol Conserv 206:37–46. https://doi.org/10.1016/j.biocon.2016.12.010
Shi W, Ayub Q, Vermeulen M, Shao RG, Zuniga S, van der Gaag K, de Knijff P, Kayser M, Xue Y, Tyler-Smith C (2009) A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP–CEPH populations. Mol Biol Evol 27:385–393. https://doi.org/10.1093/molbev/msp243
Skrbinšek T, Jelenčič M, Waits L, Kos I, Jerina K, Trontelj P (2012) Monitoring the effective population size of a brown bear (Ursus arctos) population using new single-sample approaches. Mol Ecol 21:862–875. https://doi.org/10.1111/j.1365-294X.2011.05423.x
Steinig EJ, Neuditschko M, Khatkar MS, Raadsma HW, Zenger KR (2016) Netview p: a network visualization tool to unravel complex population structure using genome-wide SNPs. Mol Ecol Resour 16:216–227. https://doi.org/10.1111/1755-0998.12442
Stoffel MA, Esser M, Kardos M, Humble E, Nichols H, David P, Hoffman JI (2016) inbreedR: an R package for the analysis of inbreeding based on genetic markers. Methods Ecol Evol 7:1331–1339. https://doi.org/10.1111/2041-210X.12588
Stuart SN, Chanson JS, Cox NA, Young BE, Rodrigues ASL, Fischman DL, Waller RW (2004) Status and trends of amphibian declines and extinctions worldwide. Science. https://doi.org/10.1126/science.1103538
Turner TF, Salter LA, Gold JR (2001) Temporal-method estimates of ne from highly polymorphic loci. Conserv Genet 2:297–308. https://doi.org/10.1023/A:1012538611944
Vandergast AG, Wood DA, Thompson AR, Fisher M, Barrows CW, Grant TJ (2016) Drifting to oblivion? Rapid genetic differentiation in an endangered lizard following habitat fragmentation and drought. Divers Distrib 22:344–357. https://doi.org/10.1111/ddi.12398
Wang J (2011) COANCESTRY: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resour 11:141–145. https://doi.org/10.1111/j.1755-0998.2010.02885.x
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:244–262. https://doi.org/10.1111/j.1752-4571.2009.00104.x
Waples RS, Luikart G, Faulkner JR, Tallmon DA (2013) Simple life-history traits explain key effective population size ratios across diverse taxa. P Roy Soc B-Biol Sci 280:20131339. https://doi.org/10.1098/rspb.2013.1339
Waples RK, Larson WA, Waples RS (2016) Estimating contemporary effective population size in non-model species using linkage disequilibrium across thousands of loci. Heredity 117:233. https://doi.org/10.1038/hdy.2016.60
Willing EM, Dreyer C, Van Oosterhout C (2012) Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 7:e42649. https://doi.org/10.1371/journal.pone.0042649
Acknowledgements
This research was funded by grants from the Australian Research Council, the Australian Biological Resources Study, the Mohamed bin Zayed Species Conservation Fund, and the Wet Tropics Management Authority. We thank the Kuranda EnviroCare community group for their assistance over the years, and have great respect for their conservation efforts for this species. In particular, we thank Cathy Retter for her tireless efforts and hospitality. Many landholders have provided access to sites and have become involved in frog conservation, and we acknowledge all of them. We thank the following people for assistance in the field: Claire Larroux, Jacque Milton, Eleanor Hoskin and Edward Bell for assistance in the field; and Craig Moritz, Keith McDonald and Alistair Freeman for assistance and advice over the years. Finally, we acknowledge the assistance of Andrzej Kilian and the team at Diversity Arrays Technology Pty Ltd.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This research was funded by an Australian Research Council APD (DP0773537 to CJH; and DP0986013 to MH) and Australian Research Council DECRA (DE130100218 to MH), an Australian Biological Resources Study Fellowship (BBR210-27 to CJH), a Mohamed bin Zayed grant (12254136), and a Wet Tropics Management Authority Grant (922).
Author information
Authors and Affiliations
Contributions
LVB performed data analyses and drafted the manuscript. CJH and MH designed the study, collected tissue samples, assisted with analyses, and drafted the manuscript. CJH secured the funding for the fieldwork, and MH and CJH secured the funding for the sequencing. KZ assisted with analyses and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare that are relevant to the content of this article.
Ethical approval
Research was conducted under Queensland Government permits (WITK04867007, WITK10437611, WISP04867207, WISP10437711), and animal ethics from the Australian National University (F.BTZ.18.07) and James Cook University (A1669, A1966, A2349).
Consent to participate
No human subjects were involved in this research.
Consent for publication
All authors consent to publication.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bertola, L.V., Higgie, M., Zenger, K.R. et al. Conservation genomics reveals fine-scale population structuring and recent declines in the Critically Endangered Australian Kuranda Treefrog. Conserv Genet 24, 249–264 (2023). https://doi.org/10.1007/s10592-022-01499-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-022-01499-7