Abstract
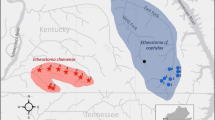
The Nashville crayfish (Faxonius shoupi, Hobbs 1948) was federally listed as an endangered species in 1986 due to its limited distribution in the Mill Creek watershed; this waterway lies in the rapidly developing Nashville basin and has experienced habitat degradation due to agricultural run-off, contamination, and urban development. Recovery efforts, including dam removal and restoration of riparian zones, have improved conditions in Mill Creek and F. shoupi has increased in numbers and recolonized extirpated stream segments. However, a history of demographic bottlenecks and restricted gene flow may have negatively impacted the long-term recovery of this species. A recently discovered population of F. shoupi in a disjunct segment of the Lower Tennessee River at the Pickwick Tailwater may provide an additional source of genetic variation. Uncertainty surrounding the origins of the Pickwick population and its taxonomic relationship to F. shoupi in Mill Creek raises questions about the conservation and management implications of this population. We used mitochondrial sequencing and SNP genotyping to assess genetic variation and connectivity of F. shoupi in the Mill Creek drainage and to investigate the taxonomy and demographic history of the newly discovered population at Pickwick. We found substantial genetic variation and evidence of connectivity for samples throughout Mill Creek for both mitochondrial and genome-wide SNPs. Our results also suggest a recently severed connection between crayfish in Pickwick and Mill Creek. Unique mitochondrial haplotypes and SNP variation in the Pickwick population highlight the need for prioritizing this population in future conservation and management plans for this species.







Similar content being viewed by others
Data Availability
Raw sequence reads obtained from GBS sequencing are available at NIH Sequence Read Archive: BioProject PRJNA817058. Mitochondrial sequences from Sanger sequencing are deposited on Genbank: Accession Numbers ON025677-ON025783.
References
Allendorf FW (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5:181–190
Barrociere LJ (1986) The ecological assessment and distribution status of the Nashville Crayfish, Orconectes shoupi. Thesis, Tennessee Technological University
Bizwell EA, Mattingly HT (2010) Aggressive Interactions of the Endangered Nashville Crayfish, Orconectes shoupi. Southeast Nat 9:359–372
Bouchard RW (1972) A contribution to the knowledge of Tennessee crayfish. Dissertation, University of Tennessee, Knoxville
Bouchard RW (1974) Crayfishes of the Nashville Basin, Tennessee, Alabama and Kentucky (Decapoda, Astacidae). Assoc Southeast Biol Bull 21:41
Bouchard RW (1976) Geography and ecology of crayfishes of the Cumberland Plateau and Cumberland Mountains, Kentucky, Virginia, Tennessee, Georgia, and Alabama. Part I. The genera of Procambarus and Orconectes. Freshw Crayfish 2:563–584
Bouchard RW (1984) Distribution and status of the endangered crayfish Orconectes shoupi (Decapoda: Cambaridae). Report, USFWS
Carpenter J (2002) Recolonization of depleted areas by the Nashville Crayfish. Report, USFWS
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4(1):s13742–s14015
Cornuet JM, Gordon L (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Couch ZL, Schuster GA (2020) Changes to Kentucky’s Crayfish Fauna Since 2004 with an Updated Checklist of Kentucky Crayfish Species. J Ky Acad Sci 80:38–46
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Durand JB (1960) Limb regeneration and endocrine activity in the crayfish. Biol Bull 118:250–261
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fitzpatrick BM, Fordyce JA, Niemiller ML, Reynolds RG (2012) What can DNA tell us about biological invasions? Biol Invasions 14:245–253
Fleming RS (1939) The larger crustacea of the Nashville region. J Tennessee Acad Sci 13:261–264
Flouri T, Jiao X, Rannala B, Yang Z (2020) A Bayesian implementation of the multispecies coalescent model with introgression for phylogenomic analysis. Mol Biol Evol 37:1211–1223
Folmer O, Hoeh WR, Black MB, Vrijenhoek RC (1994) Conserved primers for PCR amplification of mitochondrial DNA from different invertebrate phyla. Mol Mar Biol Biotech 3:294–299
Galloway WE, Whiteaker TL, Ganey-Curry P (2011) History of Cenozoic North American drainage basin evolution, sediment yields, and accumulation in the Gulf of Mexico basin. Geosphere 7:938–973
Gautier M et al (2013) The effect of RAD allele dropout on the estimation of genetic variation within and between populations. Mol Ecol 22:3165–3178
Grubb B (2020) Phylogeography and population genetics of a headwater-stream adapted crayfish, Cambarus pristinus (Decapoda: Cambaridae), from the Cumberland Plateau in Tennessee. M.S. Thesis Austin Peay State University.
Haag WR, Cicerello RR (2016) A distributional atlas of the freshwater mussels of Kentucky. Scientific and Technical Series 8, Kentucky State Nature Preserves Commission, Frankfort.
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic Acids Symposium Series, vol 41, pp 95–98
Hartl DL, Clark AG (1997) Principles of population genetics, vol 116. Sinauer Associates, Sunderland
Hartl DL, Clark AG (1997b) Principles of Population Genetics, 3rd edn. Sinauer Associates Inc, Sunderland, MA
Hasegawa M, Kishino H, Yano T (1985) Dating the human-ape split by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174
Hedgecock D, Pudovkin AI (2011) Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull Mar Sci 87:971–1002
Hedrick P (2005) Large variance in reproductive success and the Ne/N ratio. Evolution 59:1596–1599
Hobbs HH Jr (1948) A new crayfish of the genus Orconectes from southern Tennessee. Proc Biol Soc Wash 61:85–92
Hobbs HH Jr (1969) On the distribution and phylogeny of the crayfish genus Cambarus. The distributional history of the biota of the southern Appalachians Part I: invertebrates. Res Div Monogr 1:93–178
Hobbs HH, Shoup CS (1942) (1942) On the crayfishes collected from the Big South Fork of the Cumberland River in Tennessee during the summer of 1938. Am Midl Nat 28(3):634–643
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Johnson JR, Thomson RC, Micheletti SJ, Shaffer HB (2011) The origin of tiger salamander (Ambystoma tigrinum) populations in California, Oregon, and Nevada: introductions or relicts? Conserv Genet 12:355–370
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genomics 11:94
Jp VÄHÄ, Erkinaro J, Niemela E, Primmer CR (2007) Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol Ecol 16:2638–2654
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Larson ER, Olden JD (2016) Field sampling techniques for crayfish. In: Longshaw M, Stebbing P (eds) Biology and ecology of crayfish. CRC Press, Boca Raton, pp 284–358
Larson ER, Abbott CL, Usio N, Azuma N, Wood KA, Herborg LM, Olden JD (2012) The signal crayfish is not a single species: cryptic diversity and invasions in the Pacific Northwest range of Pacifastacus leniusculus. Freshw Biol 57:1823–1838
Loiseau MT, Pudlo P, Kerdelhué C, Estoup A (2013) Estimation of population allele frequencies from next-generation sequencing data: pool-versus individual-based genotyping. Mol Ecol 22:3766–3779
Luikart G, Allendorf FW, Cornuet JM, Sherwin WB (1998) Distortion of allele frequency distributions provides a test for recent population bottlenecks. J Hered 89:238–247
Meirmans PG (2006) Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60:2399–2402
Meirmans PG, Hedrick PW (2011) Assessing population structure: FST and related measures. Mol Ecol Resour 11:5–18
Miller AC, Hartheld PD, Rhodes L (1990) An investigaion of Orconectes shoupi in Mill Creek and Sevenmile Creeks, Tennessee. J Tennessee Acad Sci 65:21–24
Miller AC, Hartfield PD (1987) A study of Orconectes shoupi, Mill Creek Basin, Tennessee, 1985. Mississippi Museum of Natural Science Jackson, Mississippi
Miller MA, Pfeiffer W, Schwartz T (2011) The CIPRES science gateway: a community resource for phylogenetic analyses. In: Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, pp 1–8
Mlinarec J, Porupski I, Maguire I, Klobučar G (2016) Comparative karyotype investigations in the white-clawed crayfish Austropotamobius pallipes species complex and stone crayfish A. torrentium (Decapoda: Astacidae). J Crustac Biol 36:87–93
Morin PA, Martien KK, Taylor BL (2009) Assessing statistical power of SNPs for population structure and conservation studies. Mol Ecol Resour 9:66–73
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 9:1–10
O'Bara CJ, Korgi AJ, Stark 0J (1985) Status survey of Nashville crayfish, Orconectes shoupi. Letter report submitted to the U.S. Fish and Wildlife Service, Asheville, North Carolina, Tennessee Technological University, Cookeville, Tennessee
Ortmann AE (1918) The naiads (freshwater mussels) of the upper Tennessee drainage. With notes on synonymy and distribution. Proc Am Philos Soc 57:521–626
Paradis E (2010) pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26:419–420
Piry S, Luikart G, Cornuet JM (1999) Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J Hered 90:502–503
Poly WJ, Wetzel JE (2003) Distribution and taxonomy of three species of Orconectes (Decapoda: Cambaridae) in Illinois, USA. J Crustac Biol 23:380–390
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Racine JS (2012) RStudio: a platform-independent IDE for R and Sweave. 167–172.
Ramasamy RK, Ramasamy S, Bindroo BB, Naik VG (2014) STRUCTURE PLOT: a program for drawing elegant STRUCTURE bar plots in user friendly interface. Springerplus 3:1–3
Rannala B, Yang Z (2017) Efficient Bayesian species tree inference under the multispecies coalescent. Syst Biol 66:823–842
Reynolds J, Souty-Grosset C, Richardson A (2013) Ecological roles of crayfish in freshwater and terrestrial habitats. Freshw Crayfish 19:197–218
Schuster GA, Taylor CA, Johansen J (2008) An annotated checklist and preliminary designation of drainage distributions of the crayfishes of Alabama. Southeast Nat 7:493–504
Silliman K, Indorf J, Knowlton N, Browne B, Hurt C (2021) Base-substitution mutation rate across the nuclear genome of Alpheus snapping shrimp and the timing of isolation by the Isthmus of Panama. BMC Ecol Evol 21(1):1–4
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504
Taylor CA, Knouft JH (2006) Historical influences on genital morphology among sympatric species: gonopod evolution and reproductive isolation in the crayfish genus Orconectes (Cambaridae). Biol J Lin Soc 89:1–12
Thoma RF, Fetzner Jr. JW (2008) Taxonomic status of Cambarus (Jugicambarus) jezerinaci, spiny scale crayfish (Powell River crayfish). Report, Virginia Department of Game & Inland Fisheries Richmond, Virginia
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
US Fish and Wildlife Service (1986) Endangered and threatened wildlife and plants; determination of endangered species status for the Nashville Crayfish (Orconectes shoupi). Federal Registrar 51:34410–34412
US Fish and Wildlife Service (1987) Nashville crayfish recovery plan. US Fish and Wildlife Service, Atlanta
US Fish and Wildlife Service (2018) Species Status Assessment Report for the Nashville Crayfish (Orconectes shoupi). US Fish and Wildlife Service, Atlanta
Waples RS (2016) Making sense of genetic estimates of effective population size. Mol Ecol 25:4689–4691
Waples RS, Do CH (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756
Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Wetzel JE, Poly WJ, Fetzner JW Jr (2004) Morphological and genetic comparisons of golden crayfish, Orconectes luteus, and rusty crayfish, O. rusticus, with range corrections in Iowa and Minnesota. J Crustac Biol 24:603–617
Willing EM, Dreyer C, Van Oosterhout C (2012) Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 7:e42649
Wright S (1938) Size of population and breeding structure in relation to evolution. Science 87:430–431
Xie W, Lewis PO, Fan Y, Kuo L, Chen MH (2011) Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol 60:150–160
Yang Z (2015) The BPP program for species tree estimation and species delimitation. Curr Zool 61:854–865
Yi S, Li Y, Shi L, Zhang L, Li Q, Chen J (2018) Characterization of population genetic structure of red swamp crayfish, Procambarus clarkii, in China. Sci Rep 8:1–11
Acknowledgements
We thank Philip Hedrick, Jeff Simmons, David Withers and Gerry Dinkins for comments and suggestions on earlier versions of the manuscript. We would also like to thank the USFWS, Edward Lisic and the URECA Team grant program for providing partial funding for this work.
Funding
Funding for this project was provided by the USFWS and through the Undergraduate Research and Creative Activities Program (URECA) at Tennessee Tech University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
N/A.
Ethical approval
N/A.
Consent to participate
N/A.
Consent for publication
N/A.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hurt, C., Hildreth, P. & Williams, C. A genomic perspective on the conservation status of the endangered Nashville crayfish (Faxonius shoupi). Conserv Genet 23, 589–604 (2022). https://doi.org/10.1007/s10592-022-01438-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-022-01438-6