Abstract
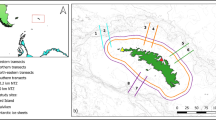
Marine protected areas (MPAs) have the potential to conserve biodiversity and improve fishery sustainability, but their efficacy depends on sound design and implementation, which requires an understanding of connectivity among reserves and between reserves and fished areas. Most studies of connectivity involving reserves focus on fishes with characteristics atypical for exploited species, making the results less applicable to fisheries management. Here, patterns of genomic diversity were assessed within and among geographic samples of juvenile of silk snapper, Lutjanus vivanus, collected in protected and fished areas on the western coast of Puerto Rico. The results indicate significant variation in spatiotemporal genetic recruitment patterns, with the two MPAs located off the shelf having partially decoupled recruitment processes from sites on the shelf. Spatial autocorrelation was found at distances less than 20 km within years, but the degree and pattern of spatial structure differed across years, suggesting that recruitment along the west coast of Puerto Rico originates from semi-independent units of spawners whose contribution varies in space and time. The results suggest that while MPAs may work to supplement fisheries where recruitment is spatiotemporally predictable, in species for which adult contribution is variable in space and time, other management strategies should be explored as well.



Similar content being viewed by others
Data availability
Sequence reads for each species have been deposited with NCBI Short Read Archive with accession number PRJNA800815. Scripts to process the sequence data are available at: https://github.com/jpuritz; https://github.com/stuartwillis; https://github.com/chollenbeck.
References
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5:384–387
Almany GR, Hamilton RJ, Matawai M, Bode M, Potuku T, Saenz-Aguadelo P, Planes S, Berumen ML, Rhodes KL, Thorrold SR et al (2013) Dispersal of grouper larvae drives local resource sharing in a coral reef fishery. Curr Biol 23:626–630
Anderson WD (2003) Lutjanidae. In: Carpenter KE (ed) The living marine resources of the western central Atlantic. pp 1479–1504
Baetscher DS, Clemento A, Ng TC, Anderson EC, Garza JC (2018) Microhaplotypes provide increased power from short-read DNA sequences for relationship inference. Mol Ecol Resour 18:296–305
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw. https://doi.org/10.18637/jss.v067.i01
Baums IB, Paris CB, Chérubin LM (2006) A bio-oceanographic filter to larval dispersal in a reef-building coral. Limnol Oceanogr 51:1969–1981
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300
Bernardi G, Beldade R, Holbrook SJ, Schmitt RJ (2012) Full-sibs in cohorts of newly settled coral reef fishes. PLoS ONE 7:e44953
Berumen ML, Almany GR, Planes S, Jones GP, Saenz-Aguadelo P, Thorrold SR (2012) Persistence of self-recruitment and patterns of larval connectivity in a marine protected area network. Ecol Evol 2:444–452
Boardman C, Weiler D (1979) Aspects of the life history of three deep water snappers around Puerto Rico, vol 32. Gulf and Caribbean Fisheries Institute, Marathon, pp 158–172
Botsford LW, Hastings A, Gaines SD (2001) Dependence of sustainability on the configuration of marine reserves and larval dispersal distance. Ecol Lett 4:144–150
Brander LM, Beukering P, Nijsten L, McVittie A, Baulcomb C, Eppink FV, Amrit J, van der Lelij C (2020) The global costs and benefits of expanding Marine Protected Areas. Marine Policy 116:103953
Broquet T, Viard F, Yearsley J (2013) Genetic drift and collective dispersal can result in chaotic genetic patchiness. Evolution 67:1660–1675
Burns KM, Brown-Peterson NJ, Overstreet RM, Gannon JG, Sprinkel JM, Robbins BD, Weaver CA (2006) Geographic comparison of age, growth, reproduction, movement, and survival ofred snapper offthe state ofFlorida. Mote Marine Laboratory Technical Report
Cabral RB, Bradley D, Mayorga J, Goodell W, Friedlander AM, Sala E, Costello C, Gaines SD (2020) A global network of marine protected areas for food. Proc Natl Acad Sci USA 117:28134–28139
Carter J, Perrine D (1994) A spawning aggregation of dog snapper, Lutjanus Jocu (Pisces: Lutjanidae) in Belize, Central America. Bull Mar Sci 55:228–234
CFMC (2013) Fact sheet: annual catch limits for US Caribbean species. F. S. 2. CFMC, San Juan
Chaytor JD, ten Brink US (2010) Extension in mona passage, Northeast Caribbean. Tectonophysics 493:74–92
Christie MR, Tissot BN, Albins MA, Beets JP, Jia Y, Ortiz DM, Thompson SE, Hixon MA (2010) Larval connectivity in an effective network of marine protected areas. PLoS ONE 5:e15715
Claro R, Lindeman KC (2003) Spawning aggregation sites of snapper and grouper species (Lutjanidae and Serranidae) on the insular shelf of Cuba. Gulf Caribb Res 14:91–106
Conover WJ, Iman RL (1979) On multiple-comparison procedures. Los Alamos Scientific Laboratory, New Mexico
Cowen RK, Lwiza KMM, Sponaugle S, Paris CB, Olson DB (2000) Connectivity of marine populations: open or closed? Science 287:857
Crowder LB, Lyman SJ, Figueira WF, Priddy J (2000) Source-sink population dynamics and the problem of siting marine reserves. Bull Mar Sci 66:799–820
Cummings NJ, Matos-Caraballo D (2003) Summarized reported commercial landings in Puerto Rico from 1969–2001 with specific notes on the silk snapper landings category. Sustainable Fisheries Division Contribution No. SFD 2003-0023 and Caribbean Deepwater SEDAR Data Workshop Report 4 SEDAR
Danacek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker R, Lunter G, Marth G, Sherry ST et al (2011) The variant call format and VCF tools. Bioinformatics 27:2156–2158
Deli T, Guizeni S, Ben Abdallah L, Said K, Chatti N (2020) Chaotic genetic patchiness in the pelagic teleost fish Sardina pilchardus across the Siculo-Tunisian Strait. Mar Biol Res 16:280–298
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator V2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839
Eldon B, Riquet F, Yearsley J, Jollivet D, Broquet T (2016) Current hypotheses to explain genetic chaos under the sea. Curr Zool 62:551–566
Figueira WF (2009) Connectivity or demography: defining sources and sinks in coral reef fish metapopulations. Ecol Model 220:1126–1137
Flowers JM, Schroeter SC, Burton RS (2002) The recruitment sweepstakes has many winners: genetic evidence from the sea urchin Strongylocentrotus purpuatus. Evolution 56:1445–1453
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Fox J, Weisberg S, Adler D et al (2011) Car: companion to applied regression. CRAN, p. R package version 2.1-4
Funk WC, McKay JK, Hohenlohe PA, Allendorf FW (2012) Harnessing genomics for delineating conservation units. Trends Ecol Evol 27:489–496
Gaines SD, White C, Carr MH, Palumbi S (2010) Designing marine reserve networks for both conservation and fisheries management. Proc Natl Acad Sci USA 107:18286–18293
Gell FR, Roberts CM (2003) Benefits beyond boundaries: the fishery effects of marine reserves. Trends Ecol Evol 18:448–455
Goudet J, Jombart T (2015) hierfstat: estimation and tests of hierarchical F-statistics. p. R package version 0.04-22
Green AL, Maypa AP, Almany GR et al (2015) Larval dispersal and movement patterns of coral reef fishes, and implications for marine reserve network design. Biol Rev (Camb) 90:1215–1247
Halpern BS, Warner RR (2003) Matching marine reserve design to reserve objectives. Proc R Soc Lond B 270:1871–1878
Hauser L, Carvalho GR (2008) Paradigm shifts in marine fisheires genetics: ugly hypotheses slain by beautiful facts. Fish Fish 9:333–362
Hedgecock D (1994) Does variance in reproductive success limit effective population sizes of marine organisms? In: Beaumont A (ed) Genetics and evolution of aquatic organisms. Chapman and Hall, London, pp 122–134
Heino M, Kaitala V, Ranta E, Lindstrom J (1997) Synchronous dynamics and rates of extinction in spatially structured populations. Proc R Soc Biol Sci Ser B 264:481–486
Hilborn R, Stokes K, Maquire JJ et al (2004) When can marine reserves improve fisheries management? Ocean Coast Manag 47:194–205
Hogan JD, Thiessen RJ, Sale PF, Heath DD (2012) Local retention, dispersal and fluctuating connectivity among populations of a coral reef fish. Oecologia 168:61–71
Hothorn T, Bretz F, Westfall P, Heiberger RM, Schuetzenmeister A, Scheibe S (2017) multcomp: simultaneous inference in general parametric models. CRAN, p. R package version 1.4-8
Johns E, Wilson WD, Molinari RL (1999) Direct observations of velocity and transport in the passages between the Intra-Americas Sea and the Atlantic Ocean, 1984–1996. J Geophys Res 104:25805–25820
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jones GP, Planes S, Thorrold SR (2005) Coral reef fish larvae settle close to home. Curr Biol 15:1314–1318
Jonsson PR, Hammar L, Wåhlström I, Pålsson J, Hume D, Almroth-Rosell E, Mattsson M (2021) Combining seascape connectivity with cumulative impact assessment in support of ecosystem-based marine spatial planning. J Appl Ecol 58:576–586
Kendall BE, Bjornstad ON, Bascompte J, Keitt TH, Fagan WF (2000) Dispersal, environmental correlation and spatial synchrony in population dynamics. Am Nat 155:628–636
Kershaw F, McClintock W, Andrews KR, Riet-Sapriza FG, Caballero S, Tetley MJ, Notarbartolo di Sciara G, Hoyt E, Goldberg G, Chou E et al (2021) Geospatial genetics: Integrating genetics into marine protection and spatial planning. Aquat Conserv 31:2440–2458
Kritzer JP, Sale PF (2004) Metapopulation ecology in the sea: from Levin’s model to marine ecology and fisheries science. Fish Fish 5:131–140
Krueck NC, Treml EA, Innes DJ, Ovenden JR (2020) Ocean currents and the population genetic signature of fish migrations. Ecology 101:e02967
Lagos NA, Tapia FJ, Navarrete SA, Castilla JC (2007) Spatial synchrony in the recruitment of intertidal invertebrates along the coast of central Chile. Mar Ecol Prog Ser 350:29–39
Leis JM (1987) Review of early life history of tropical groupers (Serranidae) and snapper Lutjanidae. In: Polovina JJ, Ralston S (eds) Tropical snappers and groupers: biology and fisheries management. Westview Press, Boulder, pp 189–237
Lenihan HS, Gallagher JP, Peters JR, Stier AC, Hofmeister JKK, Reed DC (2021) Evidence that spillover from Marine Protected Areas benefits the spiny lobster (Panulirus interruptus) fishery in southern California. Sci Rep 11:2663
Manni F, Guérard E, Heyer E (2004) Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by “Monmonier’s algorithm.” Hum Biol 76:173–190
Matos-Caraballo D (2008) Lessons learned from the Puerto Rico’s Commercial Fishery, 1988–2008. 61st Gulf and Caribbean Fisheries Institute. Proceedings of the 61st Gulf and Caribbean Fisheries Institute
McEachran J, Fechhelm JD (2005) Fishes of the Gulf of Mexico: scorpaeniformes to tetraodontiformes. University of Texas Press, Austin
Meirmans PG, Van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Moore JW, Connors BM, Hodgson EE (2021) Conservation risks and portfolio effects in mixed-stock fisheries. Fish Fish 22:1024–1040
Moran P (1953) The statistical analysis of the Canadian lynx cycle II. Synchronization and meteorology. Aust J Zool 1:291–298
Musick JA (1999) Ecology and conservation of long-lived marine animals. Am Fish Soc Symp 23:1–10
Myers RA, Mertz G, Bridson J (1997) Spatial scales of interannual recruitment variations of marine, anadromous, and freshwater fish. Can J Fish Aquat Sci 54:1400–1407
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
NOAA (2010) Public hearing draft amendment 2 to the fishery management plan for the queen conch fishery of Puerto Rico and the U.S. Virgin Islands and amendment 5 to the reef fish fishery management plan of Puerto Rico and the U.S. Virgin Islands (with Draft Environmental Impact Statement and Regulatory Impact Review). National Oceanographic and Atmospheric Administration
NOAA (2011) National Marine Fisheries Service 2010 report to Congress on the status of U.S. Fisheries. National Oceanographic and Atmospheric Administration
O’Leary SJ, Puritz JB, Willis SC, Hollenbeck CM, Portnoy DS (2018) These aren’t the loci you’re looking for: principles of effective SNP filtering for molecular ecologists. Mol Ecol 27:3193–3206
Oken KL, Holland DS, Punt AE (2021) The effects of population synchrony, life history, and access constraints on benefits from fishing portfolios. Ecol Appl 31:e2307
Ong JJL, Walter JA, Jensen OP, Pinsky ML (2021) Global hotspots of coherent marine fishery catches. Ecol Appl 31:e02321
Ottmann D, Grorud-Colvert K, Sard NM, Huntington BE, Banks MA, Sponaugle S (2016) Long-term aggregation of larval fish siblings during dispersal along an open coast. Proc Natl Acad Sci USA 113:14067–14072
Paris CB, Cowen RK, Claro R, Lindeman KC (2005) Larval transport pathways from Cuban snapper (Lutjanidae) spawning aggregations based on biophysical modeling. Mar Ecol Prog Ser 296:93–106
Patterson WF, Watterson JC, Shipp RL, Cowan J (2001) Movement of tagged red snapper in the Northern Gulf of Mexico. Trans Am Fish Soc 130:533–545
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HH (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135
Pew J, Wang J, Muir P, Frasier T (2015) Related: an R package for analyzing pairwise relatedness data based on codominant molecular markers. CRAN
Pittman SJ, Heyman WD (2020) Life below water: Fish spawning aggregations as bright spots for a sustainable ocean. Conserv Lett 13:e12722
Pittman SJ, Monaco ME, Friedlander AM, Legare B, Nemeth RS, Kendall MS, Poti M, Clark RD, Wedding LM, Caldow C (2014) Fish with chips: tracking reef fish movements to evaluate size and connectivity of caribbean marine protected areas. PLoS ONE 9:e96028
Pohlert T (2014) The pairwise multiple comparison of mean ranks package (PMCMR). CRAN.
Pulliam HR (1988) Souces, sinks, and population regulation. Am Nat 132:652–661
Puritz JB, Hollenbeck CM, Gold JR (2014) dDocent: a RADseq, variant-calling pipeline designed for population genomics of non-model organism. PeerJ 2:e431
Puritz JB, Gold JR, Portnoy DS (2016) Fine-scale partitioning of genomic variation among recruits in an exploited fishery: causes and consequences. Sci Rep 6:36095
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reguera-Rouzaud N, Díaz-Viloria N, Pérez-Enríquez R, Espino-Barr E, Rivera-Lucero MI, Munguía-Vega A (2021) Drivers for genetic structure at different geographic scales for Pacific red snapper (Lutjanus peru) and yellow snapper (Lutjanus argentiventris) in the tropical eastern Pacific. J Fish Biol 98:1267–1280
Roberts CM (1998) Sources, sinks, and the design of marine reserve networks. Fisheries 23:16–19
Roberts CM, Bohnsack JA, Gell FR, Hawkins JP, Goodridge R (2001) Effects of marine reserves on adjacent fisheries. Science 294:1920–1923
Rosario A, Rojas J, Piñero E, Figuerola M, Peña N, Torres W (2006) Reproductive cycle and maturation size of silk snapper (Lutjanus vivanus). Caribbean Fishery Management Council.
Rosario-Llantín J (2000) Tidal currents in mona passage. Department of Marine Sciences, University of Puerto Rico, Mayagüez
Roughgarden J, Gaines SD, Possingham H (1988) Recruitment dynamics in complex life cycles. Science 241:1460–1466
Rueger T, Harrison HB, Buston PM, Gardiner NM, Berumen ML, Jones GP (2020) Natal philopatry increases relatedness within groups of coral reef cardinalfish. Proc R Soc Biol Sci Ser B 287:20201133
Saillant E, Bradfield SC, Gold JR (2010) Genetic variation and spatial autocorrelation among young-of-the-year red snapper (Lutjanus campechanus) in the northern Gulf of Mexico. ICES J Mar Sci 67:1240–1250
Sale PF, Cowen RK, Danilowicz BS et al (2005) Critical science gaps impede use of no-take fishery reserves. Trends Ecol Evol 20:74–80
Schulzitski K, Sponaugle S, Hauff M, Walter KD, Cowen RK (2016) Encounter with mesoscale eddies enhances survival to settlement in larval coral reef fishes. Proc Natl Acad Sci USA 113:6928–6933
SEDAR (2003) Data report of SEDAR 4, Atlantic and Caribbean deepwater snapper-grouper. South Atlantic Fishery Management Council, Charleston
Seutin G, White BN, Boag PT (1991) Preservation of avian blood and tissue samples for DNA analyses. Can J Zool 69:82–90
Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82:561–573
Swearer SE, Caselle JE, Lea DW, Warner RR (1999) Larval retention and recruitment in an island population of a coral-reef ®sh. Nature 402:799–802
Sylvester JR (1974) A preliminary study of the length composition, distribution, and relative abundance of three species of deepwater snappers from the Virgin Islands. J Fish Biol 6:43–49
Sylvester JR, Drew DW, Dammann AE (1980) Selective life history of silk and blackfin snapper from the Virgin Islands. Caribb J Sci 15:41–48
Szedlmayer ST, Conti J (1999) Nursery habitats, growth rates, and seasonality of age-0 red snapper, Lutjanus campechanus, in the northeast Gulf of Mexico. Fish Bull 97:626–635
Tabash FA, Sierra LM (1996) Assessment of Lutjanus vivanus and Lutjanus buccanella in the north Caribbean coast of Costa Rica. Naga 19:48–51
Thorrold SR, Jones GP, Hellberg ME, Burton RS, Swearer SE, Neigel JE, Morgan SG, Warner RR (2002) Quantifying larval retention and connectivity in marine populations with artificial and natural markers. Bull Mar Sci 70:291–308
Vendrami DLJ, Peck LS, Clark M, Eldon B, Meredith M, Hoffman JI (2021) Sweepstake reproductive success and collective dispersal produce chaotic genetic patchiness in a broadcast spawner. Sci Adv 7:625
Wang J (2002) An estimator for pairwise relatedness using molecular markers. Genetics 160:1203–1215
Wang J (2007) Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet Res 89:135–153
Waples RS (1998) Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J Hered 89:438–450
Waples RS (2005) Genetic estimates of contemporary effective population size: to what time periods do the estimates apply? Mol Ecol 14:3335–3352
Waples RS (2006) A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7:167–184
Watson JR, Siegel DA, Kendall BE, Mitarai S, Rassweiller A, Gaines SD (2011) Identifying critical regions in small-world marine metapopulations. Proc Natl Acad Sci USA 108:E907–E913
Weir B, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Weir B, Hill WG (2002) Estimating F-statistics. Annu Rev Genet 36:721–750
White TD, Ong T, Ferretti F, Block BA, McCauley DJ, Micheli F, De Leo GA (2020) Tracking the response of industrial fishing fleets to large marine protected areas in the Pacific Ocean. Conserv Biol 34:1571–1578
Willis SC, Hollenbeck CM, Puritz JB, Gold JR, Portnoy DS (2017) Haplotyping RAD loci: an efficient method to filter paralogs and account for physical linkage. Mol Ecol Resour 17:955–965
Zapata FA, Herrón PA (2002) Pelagic larval duration and geographic distribution of tropical eastern Pacific snappers (Pisces: Lutjanidae). Mar Ecol Prog Ser 230:295–300
Acknowledgements
The authors appreciate the contributions of J. Gold, N. Cummings, D. Matos-Caraballo, M. Figuerola and F. Lentz, including helpful discussions about Puerto Rico ‘chillo’ fisheries and biooceanography. We also acknowledge the contributions of T. Krabbenhoft (U. Buffalo) to library preparation and sequencing of some individuals utilized in this study. This is publication 32 Hof the Marine Genomics Laboratory and 125 of Genetic Studies in Fishes (Genetic Studies in Marine Fishes).
Funding
National Marine Fisheries Service, National Oceanic and Atmospheric Administration (CRP Grant No. NA12NMF4540082).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10592_2021_1426_MOESM3_ESM.xlsx
Supplemental Table 1. Sampling site coordinates and individual sample length, mass, sample site, and date for the juvenile silk snapper utilized in this study. (XLSX 76 KB)
10592_2021_1426_MOESM4_ESM.xlsx
Supplemental Table 2. AMOVA-based FST for silk snapper site-years (above diagonal), and p-values from permutation (below diagonal). Abbreviations follow Table 1. Site-years with insufficient sample size have been omitted. Gray, italics: significant before correction for multiple tests; bold: significant after correction. (XLSX 55 KB)
10592_2021_1426_MOESM5_ESM.xlsx
Supplemental Table 3. AMOVA-based FST for silk snapper sites (above diagonal), and p-values from permutation (below diagonal). Abbreviations follow Table 1. Gray, italics: significant before correction for multiple tests; bold: significant after correction. (XLSX 47 KB)
10592_2021_1426_MOESM6_ESM.xlsx
Supplemental Table 4. AMOVA-based FST for juvenile silk snapper collection years and adults (above diagonal), and p-values from permutation (below diagonal). Abbreviations follow Table 1. Gray, italics: significant before correction for multiple tests; bold: significant after correction. (XLSX 37 KB)
10592_2021_1426_MOESM7_ESM.xlsx
Supplemental Table 5. Statistics of genetic diverisity for juvenile silk snapper collected at each site and year. (XLSX 54 KB)
10592_2021_1426_MOESM8_ESM.xlsx
Supplemental Table 6. Tukey post-hoc tests of mean differences in allelic richness, gene diversity, or local FST among sites of silk snapper juveniles. Abbreviations follow Table 1. Significant p-values in bold. (XLSX 61 KB)
10592_2021_1426_MOESM9_ESM.xlsx
Supplemental Table 7. Tukey post-hoc tests of mean differences in allelic richness, gene diversity, or local FST between protected and non-protected sites of silk snapper juveniles within years. (XLSX 32 KB)
10592_2021_1426_MOESM10_ESM.xlsx
Supplemental Table 8. Post-hoc Wilcoxon rank tests of mean differences in allelic richness, gene diversity, or local FST among sites of silk snapper juveniles within years. Abbreviations follow Table 1. Significant p-values in bold. (XLSX 52 KB)
10592_2021_1426_MOESM11_ESM.xlsx
Supplemental Table 9. Pairs of individuals with triadic relatedness values corresponding to relationships of named degree (FS, full sibling; HS, half sibling, avuncular, etc.), for juveniles of silk snapper. Confidence intervals were calculated by bootstrap with a moment estimator (Wang 2002). Abbreviations follow Table 1. (XLSX 50 KB)
10592_2021_1426_MOESM12_ESM.xlsx
Supplemental Table 10. Wilcoxon tests of differences in mean relatedness by classifications of relationships of silk snapper juveniles. Abbreviations follow Table 1. (XLSX 38 KB)
10592_2021_1426_MOESM13_ESM.xlsx
Supplemental Table 11. Conover-Iman tests of differences in mean relatedness between individuals within years of juvenile and adult silk snapper from the Mona Passage. On diagonal: mean relatedness for each year; below diagonal: p-value adjusted for multiple comparisons. Abbreviations follow Table 1. (XLSX 34 KB)
10592_2021_1426_MOESM14_ESM.xlsx
Supplemental Table 12. Conover-Iman tests of differences in mean relatedness between individuals within site-years of juvenile silk snapper from the Mona Passage. On diagonal: mean relatedness for each year; below diagonal: p-value adjusted for multiple comparisons. Abbreviations follow Table 1. (XLSX 44 KB)
10592_2021_1426_MOESM15_ESM.xlsx
Supplemental Table 13. Estimated effective number of breeders for silk snapper Mona Passage juveniles and adults by site-year and pooled. Site-years with insufficient sample sizes have been omitted. Abbreviations follow Table 1. (XLSX 42 KB)
10592_2021_1426_MOESM16_ESM.xlsx
Supplemental Table 14. Estimated kinship values for juvenile silk snapper collection years and adults, with p-values from Conover’s tests of differences in mean relatedness. (XLSX 36 KB)
10592_2021_1426_MOESM17_ESM.xlsx
Supplemental Table 15. Tukey post-hoc tests of mean differences in allelic richness, gene diversity, or local FST between year cohorts of silk snapper juveniles and adults. (XLSX 28 KB)
Rights and permissions
About this article
Cite this article
Willis, S.C., Hollenbeck, C.M., Puritz, J.B. et al. Genetic recruitment patterns are patchy and spatiotemporally unpredictable in a deep-water snapper (Lutjanus vivanus) sampled in fished and protected areas of western Puerto Rico. Conserv Genet 23, 435–447 (2022). https://doi.org/10.1007/s10592-021-01426-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-021-01426-2