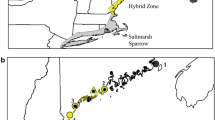
Abstract
Species range expansions and contractions can have ecological and genetic consequences, and thus are important areas of study for conservation. Hybridization and introgression are not uncommon in closely related populations that experience secondary contact during a range expansion. Allen’s Hummingbird (Selasphorus sasin) in California comprises two subspecies: the migratory S. s. sasin, which winters in central Mexico and breeds in central and northern California, and the resident S. s. sedentarius, which lives and breeds year-round on several of the Channel Islands off the California coast. Within recent decades, Allen’s Hummingbirds have been found living and breeding year-round in the southern California peri-urban mainland near Los Angeles. Ornithologists assumed that the L.A. birds were an expansion of the island subspecies, S. s. sedentarius due to similar but very subtle morphological characteristics. However, the genetic relationships among the three putative populations of Allen's hummingbird—migratory, southern California mainland, and island—are unknown. We investigated these relationships by analyzing variation of single nucleotide polymorphisms from the three geographic regions where S. sasin are present. Our population genomic analyses indicate that S. sasin hummingbirds inhabiting mainland southern California are a hybrid population resulting from admixture between S. s. sasin and S. s. sedentarius. From one perspective, these results may be interpreted as a positive development for S. s. sasin as the growing population represent an overall increase in the S. sasin population, and the expanding population contains a significant representation of S. s. sasin alleles.
Similar content being viewed by others
Data and code availability
The datasets generated during this study will be available in Dryad data repository: https://doi.org/10.5061/dryad.zgmsbcc84.
Change history
22 September 2020
In the original publication of the article, the Acknowledgements section was published incorrectly. The correct Acknowledgements section is given in this Correction.
References
Advanced Research Computing Center (2018) Teton computing environment. University of Wyoming, Laramie. https://doi.org/10.15786/M2FY47
Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001) The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16:613–622
Arnold ML (1997) Natural hybridization and evolution. Oxford University Press, New York
Arnold ML (2016) Anderson’s and Stebbins’ prophecy comes true: genetic exchange in fluctuating environments. Syst Bot 41:4–17
Avise JC, Nelson WS (1989) Molecular genetic relationships of the extinct dusky seaside sparrow. Science 243:646–648
Barton NH, Hewitt GM (1989) Adaptation, speciation and hybrid zones. Nature 341:497
Becker M, Gruenheit N, Steel M et al (2013) Hybridization may facilitate in situ survival of endemic species through periods of climate change. Nat Clim Change 3:1039
Bellard C, Thuiller W, Leroy B et al (2013) Will climate change promote future invasions? Glob Change Biol 19:3740–3748
Berthold P, Wiltschko W, Miltenberger H, Querner U (1990) Genetic transmission of migratory behavior into a nonmigratory bird population. Experientia 46:107–108
Bowlin MS, Bisson I-A, Shamoun-Baranes J et al (2010) Grand challenges in migration biology. Integr Comp Biol 50:261–279
Bradburd GS, Ralph PL, Coop GM (2016) A spatial framework for understanding population structure and admixture. PLoS Genet 12:e1005703
Charmantier A, Gienapp P (2014) Climate change and timing of avian breeding and migration: evolutionary versus plastic changes. Evol Appl 7:15–28
Chhatre VE, Emerson KJ (2017) StrAuto: automation and parallelization of STRUCTURE analysis. BMC Bioinform 18:192
Clark CJ (2017) eBird records show substantial growth of the Allen’s hummingbird (Selasphorus sasin sedentarius) population in urban Southern California. The Condor 119:122–130. https://doi.org/10.1650/CONDOR-16-153.1
Clark CJ, Mitchell DE (2013) Allen’s Hummingbird (Selasphorus sasin), version 2.0. In: Poole AF (ed) The birds of North America. Cornell Lab of Ornithology, Ithaca, NY. https://doi.org/10.2173/bna.501
Contina A, Bridge ES, Ross JD et al (2018) Examination of Clock and Adcyap1 gene variation in a neotropical migratory passerine. PLoS ONE 13:e0190859
Cooper DS (2002) Geographic associations of breeding bird distribution in an urban open space. Biol Conserv 104:205–210
De La Torre AR, Roberts DR, Aitken SN (2014) Genome-wide admixture and ecological niche modelling reveal the maintenance of species boundaries despite long history of interspecific gene flow. Mol Ecol 23:2046–2059. https://doi.org/10.1111/mec.12710
DeSante DF, George TL (1994) Population trends in the landbirds of western North America. Stud Avian Biol 15:173–190
Dusek RJ, Hall JS, Nashold SW et al (2011) Evaluation of Nobuto filter paper strips for the detection of avian influenza virus antibody in waterfowl. Avian Dis 55:674–676
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Fox D (2007) Back to the no-analog future? Science 316:823–825
Fu L, Niu B, Zhu Z et al (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Garroway CJ, Bowman J, Holloway GL et al (2011) The genetic signature of rapid range expansion by flying squirrels in response to contemporary climate warming: genetics of rapid range expansion. Glob Change Biol 17:1760–1769. https://doi.org/10.1111/j.1365-2486.2010.02384.x
Genovart M (2009) Natural hybridization and conservation. Biodivers Conserv 18:1435
Grant PR, Grant BR (1992) Hybridization of bird species. Science 256:193–197
Grinnell J, Miller AH (1944) The distribution of the birds of California. Cooper Ornithological Club, Berkeley, CA
Hamilton JA, Miller JM (2016) Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv Biol 30:33–41
Hansen AJ, Neilson RP, Dale VH et al (2001) Global change in forests: responses of species, communities, and biomes: interactions between climate change and land use are projected to cause large shifts in biodiversity. AIBS Bull 51:765–779
Hedrick PW (2013) Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol 22:4606–4618
Hinton JW, Heppenheimer E, West KM et al (2019) Geographic patterns in morphometric and genetic variation for coyote populations with emphasis on southeastern coyotes. Ecol Evol 9:3389–3404
Hobbs RJ, Higgs E, Harris JA (2009) Novel ecosystems: implications for conservation and restoration. Trends Ecol Evol 24:599–605
Hody JW, Kays R (2018) Mapping the expansion of coyotes (Canis latrans) across North and Central America. ZooKeys 759:81
Hohenlohe PA, Amish SJ, Catchen JM et al (2011) Next-generation RAD sequencing identifies thousands of SNPs for assessing hybridization between rainbow and westslope cutthroat trout. Mol Ecol Resour 11:117–122. https://doi.org/10.1111/j.1755-0998.2010.02967.x
Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589
Jetz W, Wilcove DS, Dobson AP (2007) Projected impacts of climate and land-use change on the global diversity of birds. PLoS Biol 5:e157
Keeley JE, Syphard AD (2019) Twenty-first century California, USA, wildfires: fuel-dominated vs. wind-dominated fires. Fire Ecol 15:24
Kopelman NM, Mayzel J, Jakobsson M et al (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinform 15:356
LaCava ME, Aikens EO, Megna LC et al (2019) Accuracy of de novo assembly of DNA sequences from double-digest libraries varies substantially among software. Mol Ecol Resour 20:360–370
Lande R (1988) Genetics and demography in biological conservation. Science 241:1455–1460
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. https://doi.org/10.1093/bioinformatics/btl158
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
López-Segoviano G, Bribiesca R, Arizmendi MDC (2018) The role of size and dominance in the feeding behaviour of coexisting hummingbirds. Ibis 160:283–292
Ma J, Amos CI (2012) Principal component analysis of population admixture. PLoS ONE 7:e40115
Mallet J (1995) A species definition for the modern synthesis. Trends Ecol Evol 10:294–299
Mallet J (2005) Hybridization as an invasion of the genome. Trends Ecol Evol 20:229–237
Mandeville EG, Parchman TL, McDonald DB, Buerkle CA (2015) Highly variable reproductive isolation among pairs of Catostomus species. Mol Ecol 24:1856–1872. https://doi.org/10.1111/mec.13118
Mayr E (1942) Systematics and the origin of species. Columbia University Press, New York
McCarthy EM (2006) Handbook of avian hybrids of the world. Oxford University Press, Oxford
McKinney ML (2002) Urbanization, biodiversity, and conservation. Bioscience 52:883. https://doi.org/10.1641/0006-3568(2002)052[0883:UBAC]2.0.CO;2
Muhlfeld CC, Kovach RP, Jones LA et al (2014) Invasive hybridization in a threatened species is accelerated by climate change. Nat Clim Change 4:620–624. https://doi.org/10.1038/nclimate2252
Myers BM, Rankin DT, Burns KJ, Clark CJ (2019) Behavioral and morphological evidence of an Allen’s × Rufous hummingbird (Selasphorus sasin × S. rufus) hybrid zone in southern Oregon and northern California. Auk 136:ukz049. https://doi.org/10.1093/auk/ukz049
National Audubon Society (2015) Audubon’s birds and climate change report: a primer for practitioners. National Audubon Society, New York. Contributors: Gary Langham, Justin Schuetz, Candan Soykan, Chad Wilsey, Tom Auer, Geoff LeBaron, Connie Sanchez, Trish Distler. Version 1.3.
Ottenburghs J (2019) Multispecies hybridization in birds. Avian Res 10:20. https://doi.org/10.1186/s40657-019-0159-4
Ottenburghs J, Ydenberg RC, Van Hooft P et al (2015) The Avian Hybrids Project: gathering the scientific literature on avian hybridization. Ibis 157:892–894
Owen JC (2011) Collecting, processing, and storing avian blood: a review. J Field Ornithol 82:339–354
Parchman TL, Gompert Z, Mudge J et al (2012) Genome-wide association genetics of an adaptive trait in lodgepole pine: association mapping of serotiny. Mol Ecol 21:2991–3005. https://doi.org/10.1111/j.1365-294X.2012.05513.x
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:e190
Peter BM, Slatkin M (2015) The effective founder effect in a spatially expanding population: founder effect in a spatially expanding population. Evolution 69:721–734. https://doi.org/10.1111/evo.12609
Pfennig KS, Kelly AL, Pierce AA (2016) Hybridization as a facilitator of species range expansion. Proc R Soc B 283:20161329
Phillips AR (1975) The migrations of Allen’s and other hummingbirds. The Condor 77:196–205. https://doi.org/10.2307/1365790
Pimm SL, Dollar L, Bass OL (2006) The genetic rescue of the Florida panther. Anim Conserv 9:115–122. https://doi.org/10.1111/j.1469-1795.2005.00010.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1111/j.1471-8286.2007.01758.x
Puritz JB, Hollenbeck CM, Gold JR (2014) dDocent: a RADseq, variant-calling pipeline designed for population genomics of non-model organisms. PeerJ 2:e431. https://doi.org/10.7717/peerj.431
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://R-project.org/. Accessed 21 Apr 2017
Ralston J, Lorenc L, Montes M et al (2019) Length polymorphisms at two candidate genes explain variation of migratory behaviors in blackpoll warblers (Setophaga striata). Ecol Evol 9:8840–8855
Rick TC, Sillett TS, Ghalambor CK et al (2014) Ecological change on California’s Channel Islands from the Pleistocene to the Anthropocene. Bioscience 64:680–692. https://doi.org/10.1093/biosci/biu094
Russell SM, Russell RO (2001) The North American Banders’ manual for banding hummingbirds. North American Banding Council, Point Reyes, CA
Seastedt TR, Hobbs RJ, Suding KN (2008) Management of novel ecosystems: are novel approaches required? Front Ecol Environ 6:547–553
Seehausen O (2004) Hybridization and adaptive radiation. Trends Ecol Evol 19:198–207
Seehausen O (2013) Conditions when hybridization might predispose populations for adaptive radiation. J Evol Biol 26:279–281
Shultz AJ, Baker AJ, Hill GE et al (2016) SNPs across time and space: population genomic signatures of founder events and epizootics in the House Finch (Haemorhous mexicanus). Ecol Evol 6:7475–7489. https://doi.org/10.1002/ece3.2444
Simberloff D (1996) Hybridization between native and introduced wildlife species: importance for conservation. Wildl Biol 2:143–150. https://doi.org/10.2981/wlb.1996.012
Small A (1994) California birds. Their status and distribution. IBIS Publishing Co., Vista, CA, p 342
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Todesco M, Pascual MA, Owens GL et al (2016) Hybridization and extinction. Evol Appl 9:892–908. https://doi.org/10.1111/eva.12367
Twyford A, Ennos R (2012) Next-generation hybridization and introgression. Heredity 108:179
Tylianakis JM, Didham RK, Bascompte J, Wardle DA (2008) Global change and species interactions in terrestrial ecosystems. Ecol Lett 11:1351–1363
US Fish and Wildlife Service (2008) Birds of conservation concern 2008. United States Department of Interior, Arlington, VA
Van Doren BM, Liedvogel M, Helm B (2017) Programmed and flexible: long-term Zugunruhe data highlight the many axes of variation in avian migratory behaviour. J Avian Biol 48:155–172
vonHoldt BM, Kays R, Pollinger JP, Wayne RK (2016) Admixture mapping identifies introgressed genomic regions in North American canids. Mol Ecol 25:2443–2453
vonHoldt BM, Brzeski KE, Wilcove DS, Rutledge LY (2018) Redefining the role of admixture and genomics in species conservation. Conserv Lett 11:e12371
Walther GR, Post E, Convey P, Menzel A, Parmesan C, Beebee TJC, Fromentin JM, Hoegh-Guldberg O, Bairlein F (2002) Ecological responses to recent climate change. Nature 416:389–395
Watterson G (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276
Wells S, Baptista LF (1979) Displays and morphology of an Anna × Allen Hummingbird Hybrid. Wilson Bull 91:524–532
Wells CP, Lavretsky P, Sorenson MD et al (2019) Persistence of an endangered native duck, feral mallards, and multiple hybrid swarms across the main Hawaiian Islands. Mol Ecol 28(24):5203–5216
Wilsey C, Bateman B, Taylor L, Wu JX, LeBaron G, Shepherd R, Koseff C, Friedman S, Stone R (2019) Survival by degrees: 389 bird species on the brink. National Audubon Society, New York
Wilson TS, Sleeter BM, Cameron DR (2016) Future land-use related water demand in California. Environ Res Lett 11:054018
Woltmann S, Stouffer PC, Burns CMB et al (2014) Population genetics of Seaside Sparrow (Ammodramus maritimus) subspecies along the Gulf of Mexico. PLoS ONE 9:e112739
Zink RM (2011) The evolution of avian migration. Biol J Linn Soc 104:237–250
Acknowledgements
The authors thank R. Colwell, S. Wethington, and L. Rogers for help with banding training and techniques; T. Drazenovich, L. Dalbeck, A. Vazquez, S. Wetzlich, and P. Smith technical assistance; UC Davis Museum of Wildlife and Fish Biology (I. Engilis and J. Trochet), Lindsay Wildlife Museum (A. Moresco; M. Anderson), California Animal Health and Food Safety Laboratory, California Department of Public Health Dead Bird Program, California Academy of Science Museum Vertebrate Collections, UC San Diego Wildlife Museum (K. Burns), C. Koehler, and P. Aigner for donating samples and/or expertise; C.A. Buerkle, M. Murphy, M. Dillon, and D. McDonald for guidance on analyses and project design; banding volunteers including G. Ernest-Hoar, B. Hoar, E. Graves, and S. Skalos for valuable field work assistance; M. Kusch, M. & D. Ashleigh, L. Hurley, M. Straub, T. Smith and other site hosts for their permission to study hummingbirds on their properties.
Funding
Financial support was provided by the US Fish and Wildlife Service Avian Health and Disease Grant (H.B.E.); Yolo Audubon Society (H.B.E.), Kelly Ornithology Grants (H.B.E. and B.L.G.), Meg and Bert Raynes Wildlife Fund (B.L.G.), Berry Biodiversity Center Grant (H.B.E.), University of Wyoming INBRE grant (B.L.G.), UC Davis Veterinary Genetics Laboratory (H.B.E.), UC Davis Academic Senate Grant (H.B.E.), University of Wyoming (H.B.E.), and anonymous donors (H.B.E.).
Author information
Authors and Affiliations
Contributions
BLG and HBE developed the hypothesis and design and collected samples. LAT, AE, and HBE assisted in sample collection and provided expertise. HBE supervised the research. BLG, MEFL, BM, RBG, KDG, SMLS, AE, LAT, and HBE wrote or substantially contributed to editing the paper or specific analyses. BLG analyzed the data with assistance and/or guidance from all other authors.
Corresponding author
Ethics declarations
Conflict of interest
Not applicable.
Ethical approval
All procedures conformed to the animal care and use protocols approved by the University of Wyoming, the University of California, Davis, state and federal permitting requirements. University of California protocols for animal use and care: 15387, 16977, 18605. University of Wyoming protocol for animal use and care: 20150716HE00183. Federal Bird Banding permit: 23765.
Consent for publication
All authors consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Godwin, B.L., LaCava, M.E.F., Mendelsohn, B. et al. Novel hybrid finds a peri-urban niche: Allen’s Hummingbirds in southern California. Conserv Genet 21, 989–998 (2020). https://doi.org/10.1007/s10592-020-01303-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-020-01303-4