Abstract
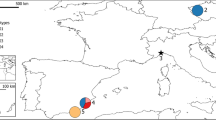
The present study investigated the natural recovery of the Eurasian otter (Lutra lutra) in France. The otter was widely distributed in France at the dawn of the 20th century, but then its range considerably shrank and became highly fragmented until the early 1970s, just before it was legally protected. However, for more than 25 years, the otter has been reconquering several parts of its original range and is now considered to be in expansion in France. We investigated the genetic differentiation and diversity of several populations from western and central France and northern Spain to gain insight into the recolonisation dynamics of this elusive species. The present study, based on the use of 14 microsatellite markers, revealed that otter populations seem to be split into five distinct groups. The distribution of samples in those five clusters was closely correlated with suspected refugia where the otter probably survived during the 20th century. Admixture was observed between genetic lineages, possibly enhancing their genetic diversity and thus increasing the recolonisation dynamics of these populations. This phenomenon resembles the genetic pattern noted in many invasive exotic species derived from multiple sources and introduction events. Finally, a demographic approach revealed the probable link between historical human pressure and otter population fragmentation patterns.






Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Ali S, Gladieux P, Leconte M, Gautier A, Justesen AF, Hovmøller MS, Enjalbert J, de Vallavieille-Pope C (2014) Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f.sp. tritici. PLoS One 10:e1003903
Arbos P, Meynier A (1936) Géographie du Massif Central. Revue de Géogr Alp 24(2):435–442
Awiti AO (2011) Biological diversity and resilience: lessons from the recovery of cichlid species in Lake Victoria. Ecol Soc 16(1):9
Bifolchi A, Picard D, Lemaire C, Cormier JP, Pagano A (2010) Evidence of admixture between differentiated genetic pools at a regional scale in an invasive carnivore. Conserv Genet 11:1–9
Bonesi L, Hale M, Macdonald DW (2013) Lessons from the use of non-invasive genetic sampling as a way to estimate Eurasian otter population size and sex ratio. Acta Theriol 58(2):157–168
Bouchardy C (1986) La loutre. Eds Le sang de la terre, Paris, p 174
Bouchardy C, Lemarchand C, Boulade Y, Gouilloux N, Rosoux R, Berny P (2011) Natural recolonisation of the Eurasian Otter (Lutra lutra) in Massif Central (France). Proc XIth Int Otter Colloquium 28B:26–29
Bryja J, Smith C, Konečný A, Reichard M (2010) Range-wide population genetic structure of the European bitterling (Rhodeus amarus) based on microsatellite and mitochondrial DNA analysis. Mol Ecol 19:4708–4722
Cabria MT, González EG, Gómez-Moliner BJ, Michaux JR, Skumatov D, Kranz A, Fournier P, Palazón S, Zardoya R (2015) Patterns of genetic variation in the endangered European mink (Mustela lutreola, 1761). BMC Evol Biol 15:141. https://doi.org/10.1186/s12862-015-0427-9
Casas-Marce M, Soriano L, López-Bao JV, Godoy JA (2013) Genetics at the verge of extinction: insights from the Iberian lynx. Mol Ecol 22:5503–5515
Chapron G, Kaczensky P, Linnell JD, Von Arx M, Huber D, Andrén H et al (2014) Recovery of large carnivores in Europe’s modern human-dominated landscapes. Science 346(6216):1517–1519
Charlesworth D, Charlesworth B (1987) Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Syst 18:237–268
Cornuet JM, Ravigné V, Estoup A (2010) Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v10). BMC Bioinf 11:401
Cornuet J-M, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A (2014) DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30(8):1187–1189
Dallas JF (1992) Estimation of microsatellite mutation rates in recombinant inbred strains of mouse. Mamm Genome 3:452–456
Dallas JF, Piertney SB (1998) Microsatellite primers for the Eurasian otter. Mol Ecol 9:1248–1251
Dallas JF, Bacon PJ, Carss DN, Conroy JWH, Green R, Jefferies DJ, Kruuk H, Marshall F, Piertney SB, Racey PA (1999) Genetic diversity in the Eurasian otter, Lutra lutra, in Scotland. Evidence from microsatellite polymorphism. Biol J Linn Soc Lond 68:73–86
Dallas JF, Marshall F, Piertney SB, Bacon PJ, Racey PA (2002) Spatially restricted gene flow and reduced microsatellite polymorphism in the Eurasian otter Lutra lutra in Britain. Genetics 3(1):15–28
Demenou BB, Pineiro RP, Hardy OJ (2016) Origin and history of the Dahomey Gap separating West and Central African rain forests: insights from the phylogeography of the legume tree Distemonanthus benthamianus. J Biogeogr 43(5):1020–1031
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genetics Resour 4(2):359–361
Edmands S (2007) Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol Ecol 16(3):463–475
Ellegren H (1995) Mutation rates at porcine microsatellite loci. Mamm Genome 6:376–377
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ferrando A, Ponsà M, Marmi J, Domingo-Roura X (2004) Eurasian Otters, Lutra lutra, have a dominant mtDNA haplotype from the Iberian Peninsula to Scandinavia. J Hered 95(5):430–435
Frankham R (2005) Resolving the genetic paradox in invasive species. Heredity 94(4):385
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24(11):2610–2618. https://doi.org/10.1111/mec.13139
Frankham R, Ballou JD, Eldridge MD, Lacy RC, Ralls K, Dudash MR, Fenster CB (2011) Predicting the probability of outbreeding depression. Conserv Biol 25:465–475
Green J, Green R (1981) The Otter (Lutra lutra L.) in western France. Mamm Rev 11:181–187
Griffiths HI, Thomas DH (1997) The conservation and management of the European badger (Meles meles). Counc Eur Nat Environ 90:86
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Janssens X, Michaux JR, Fontaine MC, Libois R, de Kermabon J, Defourny P, Baret PV (2008) Non-invasive genetics at basin scale reveals barriers to the present otter recovery in Southern France. Ecography 31:176–186
Jefferies DJ (1989) The changing otter population in Britain 1700–1989. Biol J Linn Soc Lond 38:61–69
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27(21):3070–3071
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4:782–788
Koelewijn HP, Pérez-Haro M, Jansman HAH, Boerwinkel MC, Bovenschen J, Lammertsma DR, Niewold FJJ, Kuiters AT (2010) The reintroduction of the Eurasian otter (Lutra lutra) into the Netherlands: hidden life revealed by noninvasive genetic monitoring. Conserv Genet 11:601–614
Kolbe JJ, Glor RE, Rodríguez Schettino L, Lara AC, Larson A, Losos JB (2004) Genetic variation increases during biological invasion by a Cuban lizard. Nature 431:177–181
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15(5):1179–1191
Kruuk H (2006) Otters: ecology, behaviour and conservation. Oxford Universtiy Press, Oxford. https://doi.org/10.1093/acprof:oso/9780198565871.001.0001
Kuhn R (2011) La Loutre d’Europe. In: Artois M, Haffner P, Jacques H, Moutou F, Maurin H (eds) Encyclopédie des carnivores de France. SFEPM, Bourges
Langella O (2007) Populations 1.2.30: Population genetic software (individuals or populations distances, phylogenetic trees). http://bioinformatics.org/~tryphon/populations/. Accessed 12 May 2015
Lavergne S, Molofsky J (2007) Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc Natl Acad Sci USA 104:3883–3888
Leblanc F, Dohogne R, Guischer V, Thuaire N, Bellanger C, Constant J (2005) Actualisation de la répartition de la Loutre d’Europe en Limousin (2003/2004). La Conservation de la Loutre, actes du XXVIIème Colloque Francophone de Mammalogie de la SFEPM, pp 9-28.
Legendre P, Legendre LFJ (1998) Numerical ecology. Elsevier Science, 852 pp
Léger F, Steinmetz J, Laoué E, Maillard J-F, Ruette S (2018) L’expansion du vison d’Amérique en France. Période 2000–2015. Faune Sauvage 138:23–31
Li YL, Liu JX (2018) StructureSelector: a web based software to select and visualize the optimal number of clusters using multiple methods. Mol Ecol Resour 18:176–177
Maran T, Skumatov D, Gomez A, Põdra M, Abramov AV, Dinets V (2016) Mustela lutreola. The IUCN Red List of Threatened Species 2016: e.T14018A45199861
Marcelli M, Polednik L, Polednikova K, Fusillo R (2012) Land use drivers of species re-expansion: inferring colonization dynamics in Eurasian otters. Divers Distrib 10:1001–1012
Mathias P (1933) Sur la répartition de la Loutre en France. Bull de la Soc centrale d’Aquicult et de pêche 40:73–78
Matocq M, Villablanca F (2001) Low genetic diversity in an endangered species: recent or historical pattern? Biol Cons 98(1):61–68
Michalska-Parda A, Brzezinski M, Zalewski A, Kozakiewicz M (2009) Genetic variability of feral and ranch American mink Neovison vison in Poland. Acta Theriol 54:1–10
Mercier L (2004) Bilan de la réintroduction de la Loutre Lutra lutra (Linné, 1758) en Alsace, France. Bull de la Soc d’histoire naturelle de Colmar 65:117–134
Michaux JR, Hardy OJ, Justy F, Fournier P, Kranz A, Cabria M, Davison A, Rosoux R, Libois R (2005) Conservation genetics and population history of the threatened European mink Mustela lutreola, with an emphasis on the west European population. Mol Ecol 14:2373–2388
Mondol S, Bruford MW, Ramakrishnan U (2013) Demographic loss, genetic structure and the conservation implications for Indian tigers. Proc Royal Soc B 280:20130496
Mucci N, Arrendal J, Ansorge H, Bailey M, Bodner M, Delibes M, Ferrando A, Fournier P, Fournier C, Godoy JA, Hajkova P, Hauer S, Heggberget TM, Heidecke D, Kirjavainen H, Krueger H-H, Kvaloy K, Lafontaine L, Lanszki J, Lemarchand C, Liukko U-M, Loeschcke V, Ludwig G, Madsen AB, Mercier L, Ozolins J, Paunovic M, Pertoldi C, Piriz A, Prigioni C, Santos-Reis M, Luis TS, Stjernberg T, Schmid H, Suchentrunk F, Teubner J, Tornberg R, Zinke O, Randi E (2010) Genetic diversity and landscape genetic structure of otter (Lutra lutra) populations in Europe. Conserv Genet 11:583–599
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170
Ogutu-Ohwayo R (1990) The decline in the native fishes of lakes Victoria and Kyoga (East Africa) and the impact of introduced fishes, especially Nile perch, Lates niloticus and Nile tilapia Oreochromis niloticus. Environ Biol Fishes 27:81–96
Olafsson K, Pampoulie C, Hjorleifsdottir S, Gudjonsson S, Hreggvidsson GO (2014) Present-day genetic structure of Atlantic salmon (Salmo salar) in Icelandic rivers and ice-cap retreat models. PLoS ONE 9:e86809
Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure in Canadian polar bears. Mol ecol 4(3):347–354
Palstra FP, Fraser DJ (2012) Effective/census population size ratio estimation: a compendium and appraisal. Ecol Evol 2:2357–2365
Pannell JR, Charlesworth B (1999) Neutral genetic diversity in a metapopulation with recurrent local extinction and recolonisation. Evolution 53:664–676
Penez J (2004) Les réseaux d’investissement dans le thermalisme au XIXe siècle en France. In Situ [En ligne], 4, online on 1st March 2004, consulted on 20th April 2019
Pérez-Haro M, Vinas J, Manas F, Batet A, Ruiz-Olmo J, Pla C (2005) Genetic variability in the complete mitochondrial control region of the Eurasian Otter (Lutra lutra) in the Iberian Peninsula. Biol J Linn Soc 86(4):397–403
Pigneur L-M, Etoundi E, Aldridge DC, Marescaux J, Yasuda N, Van Doninck K (2014) Genetic uniformity and long-distance clonal dispersal in the invasive androgenetic Corbicula clams. Mol Ecol 23(20):5102–5116
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Heredity 95(6):536–539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puechmaille SJ (2016) The program STRUCTURE does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16(3):608–627
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org. Accessed 20 July 2015
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Randi E, Davoli F, Pierpaoli M, Pertoldi C, Madsen AB, Loeschcke V (2003) Genetic structure in otter (Lutra lutra) populations in Europe: implications for conservation. Anim Conserv 6:1–10
Reuther C, Krekemeyer A (2004) On the way towards an Otter Habitat Network Europe (OHNE). Method and results of an assessment on the European and the German level. Habitat 15:308
Rice WR (1989) Analyzing tales of statistical tests. Evolution 43:223–225
Roman J, Darling JA (2007) Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol 22:454–464
Saavedra D, Sargatal J (1997) Reintroduction of the otter (Lutra lutra) in northeast Spain (Girona province). Galemys 10:191–199
Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O’Neil P, Parker IM, Thompson JN, Weller SG (2001) The population biology of invasive species. Annu Rev Ecol Syst 32:305–332
Slatkin M (1977) Gene flow and genetic drift in a species subject to frequent local extinctions. Theor Popul Biol 12:253–262
Spielman D, Brook BW, Briscoe DA, Frankham R (2004) Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet 5(4):439–448
Stanton DWG, Hobbs GI, McCafferty DJ, Chadwick EA, Philbey AW, Saccheri IJ, Slater FM, Bruford MW (2014) Contrasting genetic structure of the Eurasian otter (Lutra lutra) across a latitudinal divide. J Mamm 95(4):814–823
Steinmetz J, Marc D, Néri F, Trichet E, Besnard A, Defos du Rau P, Bodin J (2014) Dynamique régionale de la loutre en Midi-Pyrénées. Faune Sauv 305:31–37
Sunnaker M, Busetto AG, Numminen E, Corander J, Foll M, Dessimoz C (2013) Approximate bayesian computation. PLoS Comput Biol 9:e1002803
Takezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399
Tsutsui ND, Suarez AV, Holway DA, Case TJ (2000) Reduced genetic variation and the success of an invasive species. Proc Natl Acad Sci USA 97:5948–5953
Valière N (2002) GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes 10:1471–1478
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Weber JL, Wong C (1993) Mutation of human short tandem repeats. Hum Mol Genet 2:1123–1128
Whiteley AR, Fitzpatrick SW, Funk WC, Tallmon DA (2015) Genetic rescue to the rescue. Trends Ecol Evol 30(1):42–49
Witte F, Msuku BS, Wanink JH, Seehausen O, Katunzi EFB, Goudswaard PC, Goldschmidt T (2000) Recovery of cichlid species in Lake Victoria: an examination of factors leading to differential extinction. Rev Fish Biol Fish 10:233–241
Zalewski A, Michalska-Parda A, Bartoszewicz M, Kozakiewicz M, Brzeziński M (2010) Multiple introductions determine the genetic structure of an invasive species population: American mink Neovison vison in Poland. Biol Conserv 143(6):1355–1363
Zalewski A, Michalska-Parda A, Ratkiewicz M, Kozakiewicz M, Bartoszewicz M, Brzeziński M (2011) High mitochondrial DNA diversity of an introduced alien carnivore: comparison of feral and ranch American mink Neovison vison in Poland. Divers Distrib 17:757–768
Zhang Y-Y, Zhang D-Y, Barrett SCH (2010) Genetic uniformity characterizes the invasive spread of water hyacinth (Eichhornia crassipes), a clonal aquatic plant. Mol Ecol 19:1774–1786
Acknowledgements
The authors thank all sample contributors from the different regions, and especially Sébastien Gautier (ONCFS), Ludovic Fleury and the following partners: Cistude-Nature, Charente-Nature, CEN Midi-Pyrénées, GREGE, ONCFS (a.o. Délégation Interrégionale Nord Ouest and Cellule Technique Sud Ouest), GMB, GMN, GMHL, LPO, Museum of Toulouse (MHNT), Fédération Aude Claire, Government of Navarra via the GANASA (Gestión Ambiental de Navarra), French ministry MTES and the DREAL’s, SFEPM and PNA Loutre. The authors thank the editor and three anonymous reviewers for their fruitful comments on the manuscript. We are also grateful to Prof. K. Van Doninck (University of Namur) for help in fundraising and logistic support.
Funding
LMP and JRM benefited from FRS-FNRS grants (“chargée de recherches” and “directeur de recherches”, respectively).
Author information
Authors and Affiliations
Contributions
CFC, DM, PF, NS, JRM and LMP designed research, performed analyses and wrote the paper. CFC, PF, DM, GC, XG, BL, FS, ES, JS and FUM contributed samples and helped for interpreting data and improving the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10592_2019_1199_MOESM1_ESM.png
Supplementary material 1 Principal component analysis performed on the first 10,000 simulated datasets using DIY ABC. The large yellow dot represents the observed dataset. (PNG 159 kb)
10592_2019_1199_MOESM2_ESM.pdf
Supplementary material 2 Results of the Bayesian clustering analysis with STRUCTURE software. Left: mean lnP(D) for each tested K value, right: ΔK values for each K (PDF 28 kb)
10592_2019_1199_MOESM4_ESM.xlsx
Supplementary material 4 Model specifications and prior distributions for demographic parameters (here for Scenario 5) (XLSX 30 kb)
10592_2019_1199_MOESM5_ESM.xlsx
Supplementary material 5 Performance analysis of the model choice procedure. D = proportion of cases in which the simulation-based model choice procedure selected a scenario as the most probable with non-overlapping confidence intervals of the posterior probabilities of each scenario. * Type-I or α-error rate. ° Type-II or β-error rate (1- Σ βi is used to determine the power of the model choice procedure) (XLSX 35 kb)
Rights and permissions
About this article
Cite this article
Pigneur, LM., Caublot, G., Fournier-Chambrillon, C. et al. Current genetic admixture between relictual populations might enhance the recovery of an elusive carnivore. Conserv Genet 20, 1133–1148 (2019). https://doi.org/10.1007/s10592-019-01199-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-019-01199-9