Abstract
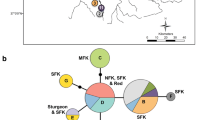
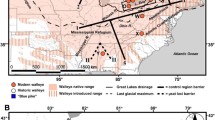
Cataloging biodiversity is of great importance given that habitat destruction has dramatically increased extinction rates. While the presence of cryptic species poses challenges for biodiversity assessment, molecular analysis has proven useful in uncovering this hidden diversity. Using nuclear microsatellite markers and mitochondrial DNA we investigated the genetic structure of Klamath speckled dace (Rhinichthys osculus klamathensis), a subspecies endemic to the Klamath–Trinity basin. Analysis of 25 sample sites within the basin uncovered cryptic diversity including three distinct genetic groups: (1) a group that is widely distributed throughout the Klamath River mainstem and its tributaries, (2) a group distributed in the Trinity River, the largest tributary to the Klamath River, and (3) a group identified above a 10 m waterfall in Jenny Creek, a small tributary to the Klamath River. All groups were resolved as divergent in nuclear microsatellite analysis and exhibited levels of divergence in mitochondrial DNA that were comparable to those observed among recognized Rhinichthys species. No physical barriers currently separate the Klamath and Trinity groups and the precise mechanism that generated and maintains the groups as distinct despite contact and hybridization is unknown. The present study highlights the importance of incorporating molecular analysis into biodiversity research to uncover cryptic diversity. We recommend that future biodiversity inventories recognize three genetically distinct groups of speckled dace in the Klamath–Trinity Basin.






Similar content being viewed by others
References
Aalto KR (2006) The Klamath peneplain: a review of JS Diller’s classic erosion surface. Geolog Soc Am Special Papers 410:451–463
Abell RM, Thieme ML, Revenga C, Bryer M, Kottelat M, Bogutskaya N, Coad B, Mandrak K, Contreras Balderas S, Bussing W, Stiassny M, Skelton P, Allen R, Unmack P, Naseka A, Ng R, Sindorf N, Robertson J, Armijo E, Higgins J, Heibel J, Wikramanake E, Olson D, Lopez H, Reis R, Lundberg J, Sabaj Perze M, Petry P (2008) Freshwater ecoregions of the world: a new map of biogeographic units for freshwater biodiversity conservation. Bioscience 58:403–414. https://doi.org/10.1641/B580507
Anderson TK (2008) Inferring bedrock uplift in the Klamath Mountains Province from river profile analysis and digital topography. Master’s thesis, Texas Tech University
Ardren WR, Baumsteiger J, Allen CS (2010) Genetic analysis and uncertain taxonomic status of threatened Foskett Spring speckled dace. Conserv Genet 11:1299–1315. https://doi.org/10.1007/s10592-009-9959-0
Avise JC (2004) Molecular markers, natural history and evolution. Springer Science & Business Media, Chicago
Baerwald MR, May B (2004) Characterization of microsatellite loci for five members of the minnow family Cyprinidae found in the Sacramento–San Joaquin Delta and its tributaries. Mol Ecol Notes 4:385–390. https://doi.org/10.1111/j.1471-8286.2004.00661.x
Baldwin EM (1981) Geology of Oregon. Kendall/Hunt Publishing, Iowa
Ballard JWO, Whitlock MC (2004) The incomplete natural history of mitochondria. Mol Ecol 13:729–744. https://doi.org/10.1046/j.1365-294X.2003.02063.x
Baltz DM, Moyle PB, Knight NJ (1982) Competitive interactions between benthic stream fishes, riffle sculpin, Cottus gulosus, and speckled dace, Rhinichthys osculus. Can J Fish Aquat Sci 39:1502–1511. https://doi.org/10.1139/f82-202
Bernardo J (2011) A critical appraisal of the meaning and diagnosability of cryptic evolutionary diversity, and its implications for conservation in the face of climate change: the systematics association, Cambridge University Press, Cambridge
Bickford D, Lohman DJ, Sodhi NS, Ng P, Meier R, Winker K, Ingram K, Das I (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22:148–155. https://doi.org/10.1016/j.tree.2006.11.004
Billman EJ, Lee JB, Young DO, McKelll M, Evans R, Shiozawa D (2010) Phylogenetic divergence in a desert fish: differentiation of speckled dace within the bonneville, lahontan, and upper snake river basins. Western North Am Nat 70:39–47
Bond CE (1994) Keys to Oregon freshwater fishes. OSU Book Stores, Corvallis
Brown LR, Moyle PB (1997) Invading species in the Eel River, California: successes, failures, and relationships with resident species. Environ Biol Fishes 49:271–291. https://doi.org/10.1023/A:1007381027518
Buggs RJA (2007) Empirical study of hybrid zone movement. Heredity 99:301–312
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach. Springer, Berlin
Carstens BC, Knowles LL (2007) Estimating species phylogeny from gene-tree probabilities despite incomplete lineage sorting: an example from Melanoplus grasshoppers. Syst Biol 56:400–411. https://doi.org/10.1080/10635150701405560
Castric V, Bonney F, Bernatchez L (2001) Landscape Structure and Hierarchical Genetic diversity in the brook charr Salvelinus Fontinalis. Evolution 55:1016–1028. https://doi.org/10.1111/j.0014-3820.2001.tb00618.x
Chapin FS III, Zavaleta ES, Eviner VT et al (2000) Consequences of changing biodiversity. Nature 405:234–242. https://doi.org/10.1038/35012241
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Dowling TE, Naylor GJ (1997) Evolutionary relationships of minnows in the genus Luxilus (Teleostei: Cyprinidae) as determined from cytochrome b sequences. Copeia 1997(4)758–765
Dowling TE, Tibbets CA, Minckley WL, Smith GR, McEachran JD (2002) Evolutionary relationships of the plagopterins (Teleostei: Cyprinidae) from cytochrome b sequences. Copeia 2002:665–678
Dudgeon D, Arthington AH, Gessner MO et al (2006) Freshwater biodiversity: importance, threats, status and conservation challenges. Biol Rev 81:163–182. https://doi.org/10.1017/S1464793105006950
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformatics 1:47–50
Felsenstein J (2005) Phylip (phylogeny inference package) version 3.6 (2004) distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Girard P, Angers B (2006) Characterization of microsatellite loci in longnose dace (Rhinichthys cataractae) and interspecific amplification in five other Leuciscinae species. Mol Ecol Notes 6:69–71. https://doi.org/10.1111/j.1471-8286.2005.01140.x
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86(6):485–486
Harrison RG (1990) Hybrid zones: Windows on evolutionary process, vol 7. In: Futuyma D, Antonovics J (ed) Oxford surveys in evolutionary biology. Oxford University Press, Oxford, pp 69–128
Harvey BC, White JL, Nakamoto RJ (2004) An emergent multiple predator effect may enhance biotic resistance in a stream fish assemblage. Ecology 85:127–133. https://doi.org/10.1890/03-3018
Heled J, Drummond AJ (2010) Bayesian inference of species trees from multilocus data. Mol Biol Evol 27:570–580
Hershey OH (1900) Ancient alpine glaciers of the Sierra Costa Mountains in California. J Geol 8:42–57
Hoekzema K, Sidlauskas BL (2014) Molecular phylogenetics and microsatellite analysis reveal cryptic species of speckled dace (Cyprinidae: Rhinichthys osculus) in Oregon’s Great Basin. Mol Phylogenet Evol 77:238–250. https://doi.org/10.1016/j.ympev.2014.04.027
Hohler DB (1981) A dwarfed population of Catostomus rimiculus (Catostomidae: Pisces) in Jenny Creek, Jackson, County, Oregon. MS Thesis, Oregon State University
Houston DD, Evans RP, Shiozawa DK (2012) Evaluating the genetic status of a Great Basin endemic minnow: the relict dace (Relictus solitarius). Conserv Genet 13:727–742. https://doi.org/10.1007/s10592-012-0321-6
Jenkins M (2003) Prospects for biodiversity. Science 302:1175–1177. https://doi.org/10.1126/science.1088666
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BioMed Central Genet 6:13. https://doi.org/10.1186/1471-2156-6-13
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BioMed Central Genetics 11:94. https://doi.org/10.1186/1471-2156-11-94
Kalinowski ST (2005) hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x
Kinziger AP, Nakamoto RJ, Anderson EC, Harvey BC (2011) Small founding number and low genetic diversity in an introduced species exhibiting limited invasion success (speckled dace, Rhinichthys osculus). Ecol Evol 1:73–84. https://doi.org/10.1002/ece3.8
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Maddison WP, Knowles LL (2006) Inferring Phylogeny despite dncomplete lineage sorting. Syst Biol 55:21–30. https://doi.org/10.1080/10635150500354928
McNeely JA, Miller KR, Reid WV, Mittermeier RA, Werner TB (1990) Conserving the world’s biodiversity. World Conservation Union (IUCN) & World Wide Fund for Nature (WWF), Gland, Suiça & World Resources Institute (WRI), Conservation International (CI) & World Bank. EUA, Washington, DC
McPhail JD, Taylor EB (2009) Phylogeography of the longnose dace (Rhinichthys cataractae) species group in northwestern North America: the origin and evolution of the Umpqua and Millicoma dace. Can J Zool 87:491–497. https://doi.org/10.1139/Z09-036
Meirmans PG, Van Tienderen PH (2004) genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794. https://doi.org/10.1111/j.1471-8286.2004.00770.x
Minckley WL, Hendrickson DA, Bond CE (1986) Geography of western North American freshwater fishes: description and relationships to intracontinental tectonism. In: Hocutt CH, Wiley EO (eds) The zoogeography of North American freshwater fishes. Wiley, New York, pp 519–613
Moyle PB (2002) Inland fishes of California. Univ of California Press
Moyle PB, Katz JVE, Quiñones RM (2011) Rapid decline of California’s native inland fishes: a status assessment. Biol Conserv 144:2414–2423. https://doi.org/10.1016/j.biocon.2011.06.002
Moyle PB, Quiñones RM, Katz JV, Weaver J (2015) Fish species of special concern in California. California Department of Fish and Wildlife, Sacramento
Murphy NP, King RA, Delean S (2015) Species, ESUs or populations? Delimiting and describing morphologically cryptic diversity in Australian desert spring amphipods. Invertebrate Syst 29:457–467. https://doi.org/10.1071/IS14036
Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858. https://doi.org/10.1038/35002501
Narum SR (2006) Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv Genet 7:783–787. https://doi.org/10.1007/s10592-005-9056-y
Niemiller ML, Graening GO, Fenolio DB et al (2013) Doomed before they are described? The need for conservation assessments of cryptic species complexes using an amblyopsid cavefish (Amblyopsidae: Typhlichthys) as a case study. Biodivers Conserv 22:1799–1820. https://doi.org/10.1007/s10531-013-0514-4
Noss RF (1990) Indicators for monitoring biodiversity: a hierarchical approach. Conserv Biol 4:355–364
Oakey DD, Douglas ME, Douglas MR (2004) Small fish in a large landscape: diversification of Rhinichthys osculus (Cyprinidae) in Western North America. Copeia. https://doi.org/10.1643/CG-02-264R1
Pearsons TN, Li HW, Lamberti GA (1992) Influence of habitat complexity on resistance to flooding and resilience of stream fish assemblages. Trans Am Fish Soc 121:427–436
Peden AE, Hughes GW (1981) Life history notes relevant to the Canadian status of the speckled dace (Rhinichthys osculus). Syesis 14:21–31
Pfenninger M, Schwenk K (2007) Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BioMed Central Evol Biol 7:121. https://doi.org/10.1186/1471-2148-7-121
Pfrender ME, Hicks J, Lynch M (2004) Biogeographic patterns and current distribution of molecular-genetic variation among populations of speckled dace, Rhinichthys osculus (Girard). Mol Phylogenet Evol 30:490–502. https://doi.org/10.1016/S1055-7903(03)00242-2
Piggott MP, Chao NL, Beheregaray LB (2011) Three fishes in one: cryptic species in an Amazonian floodplain forest specialist. Biol J Linn Soc 102:391–403
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2016). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Ricciardi A, Rasmussen JB (1999) Extinction rates of north american freshwater fauna. Conserv Biol 13:1220–1222. https://doi.org/10.1046/j.1523-1739.1999.98380.x
Rice WR (1989) Analyzing tables of statistical tests. Evol Int J Org Evol 43:223–225
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138. https://doi.org/10.1046/j.1471-8286.2003.00566.x
Sharp RP (1960) Pleistocene glaciation in the Trinity Alps of northern California. Am J Sci 258:305–340. https://doi.org/10.2475/ajs.258.5.305
Smith GR (1973) Analysis of several hybrid Cyprinid fishes from Western North America. Copeia 1973:395–410. https://doi.org/10.2307/1443102
Smith GR, Dowling TE (2008) Correlating hydrographic events and divergence times of speckled dace (Rhinichthys, Teleostei, Cyprinidae) in the Colorado River drainage. In: Reheis M, Hershler R, Miller D (eds) Late cenozoic drainage history of the southwestern Great Basin and lower Colorado River region: Geologic and biotic perspectives. Special paper 439. Geological Society of America, Boulder, pp 301–317
Tamura K, Stecher G, Peterson D, Flipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Turner TF, Dowling TE, Broughton RE, Gold JR (2004) Variable microsatellite markers amplify across divergent lineages of cyprinid fishes (subfamily Leusicinae). Conserv Genet 5:279–281. https://doi.org/10.1023/B:COGE.0000029998.11426.ab
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Waters JM, Rowe DL, Burridge CP, Wallis GP (2010) Gene trees versus species trees: reassessing life-history evolution in a freshwater fish radiation. Syst Biol 59:504–517. https://doi.org/10.1093/sysbio/syq031
Acknowledgements
The authors would like to thank the following for help with field collections: Conrad Newell, Sam Rizza, and Robbie Mueller (Humboldt State University (HSU)), Bret Harvey (US Forest Service), Rodney Nakamoto (US Forest Service), Bill Tinniswood (Oregon Department of Fish and Wildlife). Thanks to Tom Huteson for help drafting Fig. 6, Chloe Joesten (HSU) for her help in the laboratory and to Dana Herman (HSU) for providing assistance with GIS analysis. Thanks to Stewart Reid (Western Fishes Inc.) for his assistance and collection of Rogue River speckled dace and to Thomas Dowling (Wayne State University) for supplying the Rogue River mtDNA sequences. Additionally, we would like to thank the three anonymous reviewers for their comments on previous versions of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wiesenfeld, J.C., Goodman, D.H. & Kinziger, A.P. Riverscape genetics identifies speckled dace (Rhinichthys osculus) cryptic diversity in the Klamath–Trinity Basin. Conserv Genet 19, 111–127 (2018). https://doi.org/10.1007/s10592-017-1027-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-017-1027-6