Abstract
The larvae of freshwater mussels in the order Unionoida are obligate parasites on fishes. Because adult mussels are infaunal and largely sessile, it is generally assumed that the majority of gene flow among mussel populations relies on the dispersal of larvae by their hosts. The objective of this study was to compare the genetic diversity and the degree of congruence between the population structures of two related freshwater mussels Leptodea leptodon and Leptodea fragilis and their fish host, Aplodinotus grunniens. Host specificity in parasites has been shown to result in greater congruence between the population structures of the two interacting species, and assessing the congruence of genetic structure of the endangered L. leptodon with its sister species L. fragilis and their sole host is an important step in understanding the impact of host dispersal on population structure. Analysis of microsatellite data indicated that despite its imperiled status, L. leptodon displayed greater genetic diversity than the more common L. fragilis. However, the population structures of all three species were incongruent even in the presence of substantial gene flow. Other factors such as habitat specificity may play a role in generating the differences in population structure observed. This study indicates that barriers to gene flow or lack of available host fish are not the cause of decline of the federally endangered L. leptodon, and suggests that alternative explanations should be considered.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The direct or indirect interactions between species are the basis of the emerging properties of ecosystems and the varied ecological processes that occur within them. Interspecific interactions have evolutionary importance because interacting species affect not only each other’s ecological roles but also their evolutionary trajectories (i.e. the Red Queen hypothesis) (Lively et al. 1990; Morran et al. 2011). Interactions in which two or more species undergo reciprocal evolutionary changes represent cases in which the participating species are more closely tied and are generally referred to as coevolutionary (Thompson 1999). Examples of some of these interactions include those between plants and pollinators (Grant and Grant 1965; Levin 1985), predators and prey (Brodie and Brodie 1990, 1991), and hosts and parasites (Dybdahl and Lively 1996; Gigord et al. 2001; Nason et al. 2002). Investigations into the population structure of parasites and their hosts have revealed a variety of patterns that appear to vary with such factors as dispersal ability of hosts and parasites (Blouin et al. 1995; McCoy et al. 2003), host specificity of the parasite (Johnson et al. 2002; Little et al. 2006; Dick and Patterson 2007), and the ecological requirements of the species involved (Campbell et al. 1974; Brooks et al. 2006). If the host specificity of a parasite is high, congruence between the population structures of the parasite and its host is predicted (Distel et al. 1994).
Freshwater mussels (Bivalvia: Unionoida) and their fish hosts represent an intriguing but underutilized system for studying the effects of hosts on the population structures of their parasites. Unionoid mussels are typically long-lived and gonochoristic. Male mussels release sperm into the water column, which is taken into the mantle cavity by female mussels and used to fertilize their eggs. Fertilized eggs are brooded by the females in modified portions of the gills until the larvae are mature. In the family Unionidae the larvae, termed glochidia, are parasitic, and must attach to a vertebrate host for a period of several weeks (Lefevre and Curtis 1912) in order to metmorphose into juveniles. Unionid mussels can be broadly categorized as either host generalists, which can parasitize a number of different host species, or host specialists which can only parasitize a single host species (Haag and Warren 1997). Once attached to the appropriate host, the glochidia become encysted within the fish tissue (Coker et al. 1921; Telda and; Fernando 1969). While embedded in a cyst, the mussel larva undergoes metamorphosis and develops the anatomy that allows it to become a filter-feeding juvenile mussel (Kat 1984; Roe et al. 1997). After metamorphosis is completed, the juvenile mussel ruptures the cyst and drops to the sediment to join the infaunal community.
Compared to their largely sessile adult phase, the parasitic glochidium phase is considered to be responsible for dispersal and the maintenance of gene flow between populations as well as the establishment of new populations (Watters 1992), and glochidia dispersal distances therefore depend on the mobility of the host fish. Some fish species that are known hosts of freshwater mussels have been shown to move relative short distances (Schwalb et al. 2011), whereas others can travel over 100 km (Funk 1955).
In this study, we compare the genetic structure of two freshwater mussel species, the federally endangered, Leptodea leptodon (Rafinesque 1820), its common congener, Leptodea fragilis (Rafinesque 1820) and their sole host, the freshwater drum Aplodinotus grunniens (Rafinesque 1819). Leptodea leptodon was once widely distributed across central North America, and was found in 56 rivers across 13 states within the Mississippi River Basin. At present, it is restricted to only 13 streams, and can only be consistently found in three Missouri streams including the Meramec, Bourbeuse, and Gasconade rivers (Oesch 1995; Szymanski 1998; USFWS 2010). In contrast, its sister species, L. fragilis is widely distributed, ranging from Canada south to Mexico and from the Appalachian Mountains west to South Dakota, and is generally considered to be common throughout much of its range (Burch 1975; Clarke 1981). The only known host fish for both species, the freshwater drum, (Barnhart et al. 1998) displays the widest natural distribution of any freshwater fish in North America (Lee et al. 1980). In addition, freshwater drum exhibit the potential to move substantial distances, tagged freshwater drum have been recaptured up to 104 km from their original point of capture (Funk 1955).
Our research goal is to investigate the population structure of these freshwater mussels and their host. Given that freshwater drum is the only reported host fish for L. leptodon and L. fragilis, we hypothesize that the population structure of both mussel species should be highly congruent with each other and their host. Alternatively, the gene flow patterns and congruency of genetic structure of the three species may be greatly influenced by other factors such as population size, genetic diversity, and habitat specificity. In that case, the population structure of all three species may lack congruency.
Materials and methods
Sample collection and DNA extraction
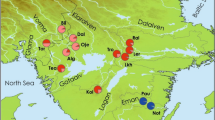
We collected samples for DNA extraction from a total of 123 L. leptodon, 104 L. fragilis, and 126 A. grunniens from the same five locations: two locations in the Gasconade River (sites A and B), one location in the Bourbeuse River (site C), and two locations in the Meramec River (sites D and E) (Table 1; Fig. 1). Samples for extraction of mussel genomic DNA were collected non-lethally by swabbing the foot and mantle, following Henley et al. (2006). Aplodinotus grunniens specimens were captured with the assistance of the Missouri Department of Conservation using a Smith-Root VI-A Electrofisher powered by a 5000-W generator in a 16-foot jon boat. After the fish were landed, a 2-mm diameter fin clip was collected and stored in 95% ETOH. Total DNA of mussel species was extracted using the Puregene Buccal Cell DNA Kit (Qiagen), and DNA was extracted from the fin clips using the Qiagen DNeasy Blood and Tissue Kit (Qiagen). The concentration of extracted genomic DNA was measured using a Nanodrop ND1000 spectrophotometer.
Sampling map for L. leptodon, L. fragilis, and A. grunniens in Missouri. Sites A and B are from Gasconade river. Site C is in Bourbeuse river. Sites D and E are in Meramec river. Genetic clusters assigned based on the LOCPRIOR model in the STRUCTURE analysis were color coded for different species: L. leptodon (green), L. fragilis (blue), and A. grunniens (yellow). (Color figure online)
Mitochondrial DNA amplification and analysis
We amplified ~ 870 base pairs (bp) of the mitochondrial ND1 gene for the two mussel species (117 L. leptodon and 100 L. fragilis) via polymerase chain reaction (PCR) using primers LEUuurF and LoglyR from Serb (2006). We amplified ~ 1100 bp of the mitochondrial cytochrome b gene from 125 A. grunniens using the Cytb F and Cytb R primers from Song et al. (1998). PCR reactions included initial denaturing at 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 52 °C for 30 s, 72 °C for 60 s, and a final extension of 72 °C for 4 min. Each PCR reaction consisted of 25 µl of reaction mix, with 12.5 µl of MyTaq (Bioline Inc.), 1 µl of 10 µM primer, 1 µl of ~ 100 ng of genomic DNA, and 9.5 µl of ddH20. PCR products for all three species were purified using the ExoSAP-IT (USB Corp.) before being bi-directionally sequenced at the Iowa State University DNA facility with an ABI 3730xl DNA Analyzer (Applied Biosystems). DNA sequences were edited using the software Geneious Pro v.5.5.6 (Drummond et al. 2010). Sequences were aligned using ClustalW as implemented in Geneious using default settings and were trimmed to equal length. We translated the nucleotide sequences to amino acids to check for alignment errors and other point mutations such as insertions/deletions. Nucleotide sequences were then grouped into haplotypes using DnaSP v.5.10.01 (Librado and Rozas 2009). The number of haplotypes, nucleotide diversity, and haplotype diversity were estimated for each population. A median-joining haplotype network was generated for each species using the programs Network v.4.613 and Network Publisher v.2.0.0.1 (Fluxus Technology, Ltd) based on optimality criteria of parsimony to infer the genetic similarity among haplotypes.
Microsatellite genotyping and analysis
Assessing genetic diversity
Ten microsatellite markers (Lele3, Lele7, Lele8, Lele9, Lele13, Lele16, Lele24, Lele30, Lele47, Lele48) from O’Bryhim et al. (2012) were used to genotype 122 L. leptodon samples. The same 10 microsatellites markers (replacing Lele16 with Lele18) were used to genotype 102 L. fragilis individuals. We optimized 11 microsatellite loci (Soc508, Soc509, Soc510, Soc521, Soc524, Soc536, Soc543, Soc551, Soc558, Soc588, Soc626) originally developed for Red drum (Karlsson et al. 2008) to genotype the 125 A. grunniens samples. PCR reactions for all loci consisted of a total of a 20 µl volume mix comprising 0.2 mm of dNTPs, 1× Biolase buffer, 1.5 of mM MgCl2, 0.25 U of Biolase Taq polymerase, 0.2 µM of M13 dye-labelled primer and non-tagged reverse primer, 0.02 µM of M13-tagged forward primer, and 15 ng template DNA. A standard annealing temperature of 55 °C was used for all PCR reactions for microsatellite loci with the following exceptions, for which a 60 °C annealing temperature was used: Lele3, Lele7, Lele8, Lele18 for L. leptodon and Lele7, Lele8, Lele18, Lele48 for L. fragilis. Scoring of the alleles was conducted using GeneMarker (Softgenetics, State College, PA).
Micro-Checker was used to assess the presence of null alleles in each marker (Van Oosterhout et al. 2004). Exact tests for Hardy–Weinberg proportions (Guo and Thompson 1992) and linkage disequilibrium were performed on all microsatellite loci using GENEPOP v.3.3 (Raymond and Rousset 1995). GenAlEx v.6.502 (Peakall and Smouse 2006, 2012) was used to calculate the number of alleles, observed and expected heterozygosity, and inbreeding coefficient for each sampling location within each species. We were also interested to know if the mussels and/or host fish populations experienced recent rapid changes in population size. A recent bottleneck may significantly alter genetic structure and connectivity among local populations, thus affecting the congruency of population structures among the mussels and their host fish. A bottleneck test was conducted on all three species on each sampling site within each species using the BOTTLENECK v.1.2 program (Piry et al. 1999) with 1000 iterations of a fixed proportion of 70% single-step model and 30% variance of geometric distribution implemented in the two-phase model.
Genetic clustering analysis and detection of migrants
Population structure was estimated for each species using STRUCTURE v.2.2 to cluster individuals into populations based on the criteria that these groups are in Hardy–Weinberg and linkage equilibrium (Pritchard et al. 2000). Two STRUCTURE analyses were conducted for each species, one with using the sample location as a prior (LOCPRIOR), and one without (Hubisz et al. 2009). An admixture model was used allowing individuals to have mixed ancestry, with 100,000 Markov chain Monte Carlo replicates following a burn-in of 50,000 runs. The correlated allele frequencies model was selected so that the refined population structure could be detected. The number of populations (K) was estimated following methods developed by Evanno et al. (2005), using STRUCTURE HARVESTER (Earl and vonHoldt 2012). A STRUCTURE bar plot was constructed using CLUMPAK (Kopelman et al. 2015). Individuals were then grouped according to the clustering results obtained from STRUCTURE. Genetic differentiation among clusters was estimated using the analysis of molecular variation (AMOVA) (Excoffier et al. 1992) to obtain pairwise F ST. F ST is known to underestimate the genetic differentiation of highly polymorphic loci (such as microsatellites) because it is based on the expected heterozygosity, which does not increase linearly when diversity increases (Jost 2008; Meirmans and Hedrick 2011). Pairwise linearized F ST (Slatkin 1995) and F’ ST values among genetic clusters were estimated with 10,000 permutations using GenAlEx v.6.502 (Peakall and Smouse 2006, 2012). However, tests of statistical significance are only presented for the pairwise linearized F ST estimates.
Individual assignment tests were estimated using GENECLASS 2 (Piry et al. 2004), where origins of individuals were considered to be correct if they were assigned to the genetic cluster identified in the STRUCTURE analysis. Individuals assigned to different clusters were taken as evidence of migration. The threshold of the assignment test was set to 5%, and a Monte Carlo resampling method of 10,000 simulations was applied following Paetkau et al. (2004). The relationship between the genetic diversity (number of alleles and observed heterozygosity) of both mussel species and their host fish were tested using linear correlation analysis, testing the null hypotheses that there is no relationship between the two variables using the t test (Zar 1984).
Congruence of inter-population divergence among species
The correlation between genetic distance and geographic distance among individuals of each species was conducted using a Mantel test (Mantel 1967) as implemented in the Isolation By Distance Web Service v.3.23 (Jensen et al. 2005). Pairwise genetic differentiation among sample sites was estimated using linearized F ST (Slatkin 1995) to construct a genetic distance matrix. Pairwise geographic distances among sites was measured using river miles in Google Earth (Google Inc.) to create a geographic distance matrix. Mantel tests were conducted with 10,000 iterations to test the relationship between genetic isolation and geographic distance (isolation by distance).
Mantel tests were also performed to examine if the genetic diversity (number of alleles and heterozygosity) and distance matrices of any pair of species were linearly correlated (α = 0.05). Because the genetic structure among populations may be influenced depending on the type of genetic distances used (Dyer et al. 2010), we conducted the Mantel tests using two types of genetic distances: linearized F ST (Slatkin 1995) and Cavalli-Sforza and Edward’s chord distances (Cavalli-Sforza and Edwards 1967). Mantel tests of the linearized F ST were performed in GenAlEx v.6.502 (Peakall and Smouse 2006) with 10,000 permutations. For the Cavalli-Sforza and Edward’s chord distances, we used the Monte Carlo test implemented in the R package “ade4” to conduct the Mantel tests, with 10,000 permutations.
Results
Leptodea leptodon mtDNA and microsatellite analysis
DNA sequencing resulted in 873 bp of sequence for the ND1 gene for 117 L. leptodon specimens. The L. leptodon mtDNA sequences were grouped into four haplotypes that were a single mutational step apart (Fig. 2). Overall nucleotide diversity of the ND1 gene was 0.0006 with moderate haplotype diversity (H d = 0.473). Two haplotypes were widespread and abundant and were observed at all sample sites. One haplotype was found only in the Meramec and Bourbeuse river populations, and the other haplotype was restricted to the upper Gasconade River populations; both of these haplotypes were observed at low frequencies (Table 2).
Mitochondrial median-joining haplotype networks constructed based on ND1 gene fragment of L. leptodon and L. fragilis, and cytochrome b gene fragment of A. grunniens from five sampling locations in Missouri. Each haplotype is represented by a node. Node size represents the number of individuals in our collection that shared the same haplotype. Black nodes (smallest node) are inferred mutational events differentiating the haplotypes. In the A. grunniens network, the number next to the interrupted lines indicates the number of mutational events greater than 4
No null alleles were detected at any microsatellite locus based on the results from Micro-Checker (Van Oosterhout et al. 2004). We compared each pair of loci and found no evidence of linkage disequilibrium (p > 0.05). Deviation from Hardy–Weinberg expectations were detected at locus Lele7 for the population from site A and locus Lele8p for the populations from sites A, B, C, and E after applying Bonferroni correction (p < 0.001). Number of alleles was similar across all sampling sites, ranging between 10 and 14 alleles per locus (Table 2). The inbreeding coefficient ranged from low (< 0.03) to intermediate (< 0.12), indicating a slight deficit in heterozygotes. The test for the occurrence of a recent bottleneck was positive in L. leptodon (p = 0.005). Genetic isolation and geographic distances were not linearly correlated in L. leptodon (r = 0.05, p = 0.46) (Fig. 3).
Isolation by distance figures for L. leptodon, L. fragilis, and A. grunniens species. Independent variable (x-axis) indicates the pairwise differences of the geographic distance (in river miles) between all sampling sites. Dependent variables (y-axis) indicates the pairwise genetic distances linearized F ST (F ST/1−F ST) among populations
The STRUCTURE analysis without using location as a prior suggested low levels of divergence and weak population structure within L. leptodon (the peak distributions of ∆K were shown in the Online Resource 1). The results from the STRUCTURE analysis using sampling sites as a prior (LOCPRIOR model) largely concurred with the STRUCTURE results without a prior. Given that the LOCPRIOR model is presumably better at detecting population structure at lower levels of divergence (Hubisz et al. 2009), in this paper we presented only the STRUCTURE results using the LOCPRIOR model.
The peak distribution of ∆K (Evanno et al. 2005) in L. leptodon STRUCTURE analysis occurred at K = 2. This result separated mussels from the Gasconade River from those in the Meramec/Bourbeuse rivers (Fig. 4). The AMOVA results (Table 3) indicate that individuals from Gasconade (sites A and B) were significantly, but weakly differentiated from individuals from the Bourbeuse and Meramec rivers (sites C, D, and E) with a linearized F ST = 0.008, p < 0.001 (F’ ST = 0.064). The GENECLASS result showed that 95% of the L. leptodon individuals were correctly assigned into the genetic clusters identified from STRUCTURE analysis (Table 4).
Leptodea fragilis mtDNA and microsatellite analysis
An 864 bp fragment of the ND1 gene was sequenced for 100 L. fragilis samples and 22 haplotypes were identified (Fig. 2). Overall nucleotide diversity was 0.0015 with high haplotype diversity (H d = 0.759). There were two common haplotypes found in all five sampling locations (Fig. 2). The remaining haplotypes occurred at low frequencies and tended to be restricted to particular sampling sites. Four rare haplotypes were unique to the Gasconade river (sites A and B); six haplotypes were restricted to Bourbeuse river (site C), and another six haplotypes were found only in the Meramec river (sites D and E).
No null alleles or linkage disequilibrium was detected in the L. fragilis microsatellite data. The number of alleles ranged from 11 to 13 for each sampling location (Table 2). After employing Bonferroni correction, significant deviations from Hardy–Weinberg expectations were detected at locus 8 for site C; locus 9 for sites A, B and C; and locus 13 for all sampling sites (p < 0.001). A test for Hardy–Weinberg equilibrium could not be performed on marker F-Lele03p due to low allelic diversity. The inbreeding coefficient was relatively similar across all sites, ranging from 0.04 to 0.08. Unlike what was observed for L. leptodon, no evidence of a recent bottleneck was detected in L. fragilis (p = 0.42). Furthermore, the Mantel test showed that the genetic distance and geographic distance were linearly correlated in L. fragilis, with r = 0.67, p = 0.046 (Fig. 3).
The STRUCTURE analysis revealed two genetic clusters (K = 2) in L. fragilis, with the upper Gasconade river (site A) forming a cluster separated from the remaining sample sites (sites B, C, D, and E) (Fig. 3). The results of the AMOVA analysis of the genotypic data also indicated that individuals from the upper Gasconade river were genetically different from individuals from the lower Gasconade, Bourbeuse, and Meramec rivers with linearized F ST = 0.038, p < 0.001 (F’ ST = 0.15) (Table 3). GENECLASS analysis indicated that the remaining 93 L. fragilis individuals from outside the upper Gasconade River region were correctly assigned to the Meramec/Bourbeuse genetic cluster. At the upper Gasconade region (site A), only 64% (4 of 11) individuals were assigned correctly to the genetic cluster identified from the STRUCTURE analysis (Table 4). Based on the results of the assignment test, genetic admixture was evident at the Gasconade river site A, where the L. fragilis population appeared to consist of a mix of individuals from both genetically distinct clusters.
Aplodinotus grunniens mtDNA and microsatellite analysis
DNA sequencing resulted in 1086 base pairs of the cytochrome b gene for 126 individual A. grunniens and 11 haplotypes were found. Overall nucleotide diversity of the mtDNA gene was 0.004. The 11 haplotypes formed two clusters that differed by seven mutational steps (Fig. 2). Three haplotypes were common and widely distributed, whereas the remaining four haplotypes occurred at low frequencies and generally were restricted to a single river basin. Two rare haplotypes were unique to the Gasconade and Meramec rivers. No rare haplotypes were documented in the Bourbeuse River.
Null alleles or linkage disequilibrium was not evident at any locus. Number of alleles among A. grunniens populations was very similar, ranging between 10 and 13 alleles (Table 2). The inbreeding coefficients were very low (F IS < 0.003) in all populations. Deviations from Hardy–Weinberg equilibrium were not detected at any locus or in any population after applying Bonferroni correction (p < 0.00091). There was no evidence of a recent bottleneck in A. grunniens (p = 0.52). Genetic isolation and geographic distance among A. grunniens were not linearly correlated in the Mantel test (r = 0.16, p = 0.32) (Fig. 3).
STRUCTURE analysis of the microsatellite data indicated K = 1, suggesting that A. grunniens from our five sampling locations belonged to a single panmictic population maintained by high gene flow and connectivity. As shown in Fig. 4, the STRUCTURE barplot of K = 2 (which looked strikingly different from the K = 2 barplots of L. leptodon and L. fragilis) clearly indicated no genetic differentiation among A. grunniens populations. Because L. leptodon and L. fragilis both showed some level of genetic differentiation between the Gasconade and Meramec/Bourbeuse populations, we conducted an AMOVA analysis by grouping the A. grunniens into these two clusters. Population differentiation analysis estimated a linearized F ST of 0.002 (F’ ST = 0.012), which was not significantly different from zero (p = 0.07), confirming the result of no population structure in A. grunniens from the Gasconade, Bourbeuse, and Meramec rivers (Table 3).
Mantel test analysis on the gene flow patterns
No correlations of genetic diversity (number of alleles and heterozygosity) between either mussel species and the host fish were found (p > 0.05) (Table 5). We conducted the Mantel tests using two types of genetic distances, and there was no correlation of the pairwise genetic distance evident using linearized F ST (F ST/1−F ST) and Cavalli-Sforza and Edward’s chord distances among populations of all three species (Table 6). In summary, the population structures of the three species are not congruent.
Discussion
In this study, we investigated patterns of genetic structure and gene flow patterns in three interacting species: two closely-related freshwater mussels (one endangered and one common) and their fish host. Our hypothesis that the host-parasite relationship between both mussels and A. grunniens would produce congruence in the population structures of all three species, was not supported by the data. The two mussel species exhibit weak population structure between river drainages that are not completely congruent, whereas the fish host displays no structure across the same area. We also observed that whereas L. fragilis and A. grunniens populations exhibit greater genetic diversity than L. leptodon populations at the mtDNA locus examined, L. leptodon displayed greater genetic diversity at microsatellite loci. The test for a bottleneck indicated that L. leptodon suffered a recent bottleneck which was not indicated using the same test in either L. fragilis or the host fish, and the reduced genetic diversity at the mtDNA locus in L. leptodon is likely a reflection of this since the effective population size of mtDNA is 1/4 that of the nuclear genome, and would be more sensitive to loss of diversity during a bottleneck. Similarly, although L. leptodon and L. fragilis have roughly the same number of alleles per locus, the expected heterozygosity of L. leptodon is higher. Expected heterozygosity responds more slowly to changes in population size than the number of alleles, and supports a population contraction for L. leptodon. In contrast to the pattern observed for L. leptodon, L. fragilis exhibits the genetic signature consistent with a population expansion; the mtDNA exhibits a star-like pattern and the expected heterozygosity is indicative of a historically smaller populaton size. (Nei et al. 1975; Maruyama and Fuerst 1984), which is in accordance with anecdotal information concerning this species.
Role of hosts in maintaining connectivity in populations of parasites
Prior investigations comparing the population structure of freshwater mussels and their host are few in number. Geist and Kuehn (2008) examined congruence between genetic diversity and population structure between the European pearl mussel (Margaritifera margaritifera) and its host, the brown trout (Salmo trutta m. fario). Even though the population structures of the two species were similar, European pearl mussels exhibited stronger differentiation between populations than brown trout (Geist and Keuhn 2008) as was also observed in this study. However, Geist and Kuehn (2008) found that measures of genetic diversity (number of alleles and heterozygosity) for mussel and host were inversely related and showed a significant negative correlation. Zanatta and Wilson (2011) conducted a comparison of mussel–host population structure and diversity for the federally endangered snuffbox mussel, Epioblasma triquetra, and its host, the common logperch, Percina caprodes. A comparison of pairwise genetic distance among sites indicated that the population structures of the mussel and fish host were largely congruent, but, no significant correlation of genetic diversity was found between the snuffbox and the logperch.
Our results are not entirely concordant with previous host–mussel co-evolution studies (Geist and Kuehn 2008; Zanatta and Wilson 2011), perhaps due to the different reproductive strategies pearl mussel, snuffbox, and L. leptodon employ to infest their host fish. All three species are considered host-specialists, utilizing either a single fish or relatively few in the case of the pearl mussel, but differences occur in how glochidia are transmitted to the host. Pearl mussels broadcast glochidia into the water column, Whereas snuffboxes capture host fish with their shells in order to infest them with glochidia (Barnhart et al. 2008). The method that mussels in the genus Leptodea use to infest host fish is not yet known. Neither species of Leptodea possess a mantle lure or any visible features to attract fish, and it has been proposed that Aplodinotus grunniens feeds on unionid bivalves, and perhaps become infested with glochidia after consuming gravid female Leptodea (Haag 2012).
Apart from mussel reproductive strategies, host dispersal capability is also varies greatly among the fish species included in these studies. Other studies comparing the genetic diversity and population structure of freshwater mussels have found genetic structuring at the watershed scale (e.g. Galbraith et al. 2015) and also noted a lack of congruence in genetic structure across the species examined. Those authors raised the possibility that observed species differences in gene flow between rivers was due to differences in host vagility (different mussel species utilizing different fish species as hosts), but such and explanation is not valid in this case as both mussels share the same host fish.
Inoue et al. (2013) noted that since freshwater mussels and their hosts live in linear river systems, mussels with vagile hosts can be dispersed across great distances which would results in low or no population differentiation. Aplodinotus grunniens, like brown trout, has higher mobility than does the common logperch. Average dispersal distance for logperch was only 30 m (Schwalb et al. 2011) while A. grunniens may travel up to 104 km (Funk 1955). Population isolation and genetic differentiation may be evident in both logperch and snuffbox populations, due to the small population size of snuffbox and the low mobility of logperch, which results in congruency of the population structures between mussels and host. In our study, L. leptodon glochidia have greater dispersal distance than that of the snuffbox because of the high mobility of host fish. However, glochidia gene flow of L. leptodon may still be significantly lower than the gene flow among A. grunniens populations because L. leptodon presumably has more isolated populations with smaller effective population size. This may explain why no correlation was found between population structures of L. leptodon and A. grunniens in contrast to snuffbox and common logperch.
Some evidence of weak population structure was observed in the two freshwater mussel species studied. In particular, L. leptodon seemed to display genetic differentiation between populations in different drainages (Gasconade vs. Meramec + Bourbeuse). The genetic and geographic distances among L. leptodon populations were not linearly correlated due to higher gene flow within the Gasconade and the Meramec/Bourbouse drainages than among drainages (these drainages are separated by segments of Missouri and Mississippi rivers). Interestingly, we didn’t observe a similar population isolation pattern in L. fragilis. Unlike L. leptodon, population differentiation was observed between L. fragilis from upper Gasconade (site A) and the remaining sites (Fig. 4). The test of isolation by distance showed that the genetic distance and geographic distance among L. fragilis populations were correlated, suggesting that L. fragilis exhibits a gene flow pattern distinct from L. leptodon. Leptodon fragilis from Gasconade site A also appear to be a mixture of individuals from two genetically distinct clusters, indicating an upstream migration of larvae. The population isolation between the upper and lower Gasconade river maybe because the movement of freshwater drum that carry L. fragilis glochidia reach the upper Gasconade river less frequently compared to other drainages, or perhaps there are less suitable habitats for L. fragilis between the upper and lower Gasconade river.
Mantel test results showed that significant correlations between population-level genetic matrices were not detected among all three species. Coevolution of host and parasites often leads to local adaptation and cospeciation (Huyse and Volckaert 2005). The ability of parasites to infest a host (infectivity) is directly linked to host-parasite interaction (Dybdahl and Storfer 2003), which depends on the spatial variation of their interactions (Thompson 1994). Often, the congruency between host and parasite evolutionary trees are due to high interactions/specificity (Johnson et al. 2002; Nieberding et al. 2004). Aplodinotus grunniens is the only known host fish for L. leptodon and L. fragilis (Barnhart et al. 1998). Aplodinotus grunniens are known for their high mobility (Funk 1955) and are common throughout the study area. Thus, the high level of gene flow observed in A. grunniens was not unexpected. Although our samples in this study only represent a small subset of the species range of A. grunniens, gene flow among the fish populations was evidently higher than gene flow among mussel populations. Leptodea fragilis is a widespread mussel species with large and more continuously distributed populations that live in the habitat where host fish is common and abundant. It may therefore have more opportunities to interact with A. grunniens, which would result in a population structure more congruent with the host fish than the endangered L. leptodon.
Our results also indicated that L. leptodon suffered a recent bottleneck that was not evident in either L. fragilis or the host fish. It is possible that the cause of the bottleneck (e.g. habitat degradation) affected all three species, but since L. leptodon is an endangered species with smaller effective population size, bottleneck would have larger impact on L. leptodon than the two other widespread species. The effect of inbreeding and genetic drift on L. leptodon populations that may be potentially more isolated (i.e., less opportunity to interact with host fish), resulted in a population structure and gene flow pattern that are not congruent with L. fragilis and freshwater drum.
Conservation implications
Our study sampled all of the known extant populations of L. leptodon, and our genetic analyses provided an overview of the population structure of this species. Comparing the genetic diversity of populations of related rare and common species of mussels has rarely been performed (Roe and Boyer 2015). In Roe and Boyer (2015) study, the rare species displayed substantially lower genetic diversity when compared to its common related species. We found lower mitochondrial genetic diversity in L. leptodon than in L. fragilis, but the opposite result was obtained for microsatellite loci for these two species. Both mussel species are dependent on their host fish for dispersal of glochidia and maintenance of gene flow. Our results indicated that the rarer L. leptodon has maintained higher nuclear gene diversity but lower population connectivity compared to its common sister taxon. Assisted gene flow will be beneficial in establishing L. leptodon populations where habitats and host fish have met the requirements for a sustainable population (Kelly and Philips 2016). A genetic rescue (Frankham 2015) to translocate L. leptodon to extirpated drainages may also be helpful in restoring L. leptodon populations to its historic range.
Habitat specificity may be a possible reason why L. fragilis is more abundant than L. leptodon, despite sharing the same host fish. Leptodea leptodon prefer gravel and sand substrates in large rivers, while L. fragilis are more tolerant of siltation of rivers and streams (Cummings and Mayer 1992). Habitat specificity may be affecting the expansion of extant populations of L. leptodon. The bottleneck that was evident in L. leptodon but not in L. fragilis or A. grunniens, is perhaps due to the fact that habitat preferred by L. leptodon has been negatively impacted by increased siltation and is less common than in the past (Szymanski 1998; Haag 2012).
Conclusions
No published examination of the genetic structure of L. leptodon and A. grunniens had been conducted prior to this project. Leptodea leptodon and L. fragilis share the same host fish and live in the same rivers in parts of Missouri. Yet, L. leptodon is critically endangered and L. fragilis is common and widely distributed. Understanding the factors threatening L. leptodon may help in understanding why some unionid mussels are more vulnerable to environmental changes. Our study revealed that the population structures and gene flow patterns of all three species were not congruent. A recent bottleneck was evident in the endangered L. leptodon but not in L. fragilis and the host fish. Future research should focus on understanding the factors implicated in the contraction of the historic range of L. leptodon. Attempts to quantify the differences in habitat preferences between these two bivalve species are recommended to aid in mussel translocation and restoration. Ecological factors that may affect the expansion of mussel population (i.e., mussel–host interactions, competition for host fish) are also worth exploring to develop appropriate conservation strategies for the endangered L. leptodon.
References
Barnhart MC, Riusech FA, Baird MS (1998) Drum is host of the scaleshell, Leptodea leptodon. Triannu Unionid Rep 16:35
Barnhart MC, Haag WR, Roston WN (2008) Adaptations to host infection and larval parasitism in Unionoida. J North Am Benthol Soc 27:370–394
Blouin MS, Yowell CA, Courtney CH, Dame JB (1995) Host movement and the genetic structure of populations of parasitic nematodes. Genetics 141:1007–1014
Brodie ED, Brodie ED (1990) Tetrodotoxin resistance in garter snakes: an evolutionary response of predators to dangerous prey. Evol Int J Org Evol 44:651–659
Brodie ED, Brodie ED (1991) Evolutionary response of predators to dangerous prey: reduction of toxicity of newts and resistance of garter snakes in island populations. Evol Int J Org Evol 45:221–224
Brooks DR, Leon-Regagnon V, McLennan DA, Zelmer D (2006) Ecological fitting as a determinant of the community structure of platyhelminth parasites of anurans. Ecology 87:S76-S85
Burch JB (1975) Freshwater unionacean clams (Mollusca: Pelecypoda) of North America. Malacological Publications, Hamburg
Campbell A, Frazer BD, Gilbert N, Gutierre AP, Mackauer M (1974) Temperature requirements of some aphids and their parasites. J Appl Ecol 11:431–438
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis models and estimation procedures. Am J Hum Genet 19:233–257
Clarke AH (1981) The freshwater molluscs of Canada. National Museum of Natural Sciences, National Museums of Canada, Ottawa
Coker RE, Shira AF, Clark HW, Howard AD (1921) Natural history and propagation of freshwater mussels. Bull US Bur Fish 37:75–181
Cummings KS, Mayer CA (1992) Field guide to freshwater mussels of the Midwest, Manual 5. Illinois Natural History Survey, Champaign, p 194
Dick CW, Patterson BD (2007) Against all odds: explaining high host specificity in dispersal-prone parasites. Int J Parasitol 37:871–876
Distel DL, Felbeck H, Cavanaugh CM (1994) Evidence for phylogenetic congruence among sulfur-oxidizing chemoautotrophic bacterial endosymbionts and their bivalve hosts. J Mol Evol 38:533–542
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2010) Geneious v55. http://wwwgeneious.com
Dybdahl MF, Lively CM (1996) The geography of coevolution: comparative population structures for a snail and its trematode parasite. Evol Int J org Evol 50:2264–2275
Dybdahl MF, Storfer A (2003) Parasite local adaptation: red queen versus suicide king. Trends Ecol Evol 18:523–530
Dyer RJ, Nason JD, Garrick RC (2010) Landscape modelling of gene flow: improved power using conditional genetic distance derived from the topology of population networks. Mol Ecol 19:3746–3759
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618
Funk JL (1955) Movement of stream fishes in Missouri. Trans Am Fish Soc 85:39–57
Galbraith HS, Zanatta DT, Wilson CC (2015) Comparative analysis of riverscape genetic structure in rare, threatened and common freshwater mussels. Conserv Genet 16(4):845–857
Geist J, Kuehn R (2008) Host-parasite interactions in oligotrophic stream ecosystems: the roles of life history strategy and ecological niche. Mol Ecol 17:997–1008
Gigord LDB, Macnair MR, Smithson A (2001) Negative frequency-dependent selection maintains a dramatic flower color polymorphism in the rewardless orchid Dactylorhiza sambucina (L) Soò. Proc Natl Acad Sci USA 98:6253–6255
Grant V, Grant KA (1965) Flower pollination in the Phlox family. Columbia University Press, New York
Guo SW, Thompson EA (1992) Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 48:361–372
Haag WR (2012) North American freshwater mussels: natural history, ecology, and conservation. Cambridge University Press, New York
Haag WR, Warren ML (1997) Host fishes and reproductive biology of 6 freshwater mussel species from the Mobile Basin, USA. J North Am Benthol Soc 16:576–585
Henley WF, Grobler PJ, Neves RJ (2006) Non-invasive method to obtain DNA from freshwater mussels (Bivalvia: Unionidae). J Shellfish Res 25:975–977
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Huyse T, Volckaert FAM (2005) Comparing host and parasite phylogenies: Gyrodactylus flatworms jumping from goby to goby. Syst Biol 54:710–718
Inoue K, Monroe EM, Elderkin CL, Berg DJ (2013) Phylogeographic and population genetic analyses reveal Pleistocene isolation followed by high gene flow in a wide ranging, but endangered, freshwater mussel. Heredity 112(3):282–290
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13 v323
Johnson KP, Williams BL, Drown DM, Adams RJ, Clayton DH (2002) The population genetics of host specificity: genetic differentiation in dove lice (Insecta: Phthiraptera). Mol Ecol 11:25–38
Jost L (2008) G(ST) and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Karlsson S, Renshaw MA, Rexroad CE III, Gold JR (2008) PCR primers for 100 microsatellites in red drum (Sciaenops ocellatus). Mol Ecol Resour 8:393–398
Kat PW (1984) Parasitism and the Unionacea (Bivalvia). Biol Rev 59:189–207
Kelly E, Phillips BL (2016) Targeted gene flow for conservation. Conserv Biol 30:259–267
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191
Lee DS, Gilbert CR, Hocutt CH, Jenkins RE, McAllister DE, Stauffer JR Jr (1980) Atlas of North American freshwater fishes. North Carolina States Museum of Natural History, Raleigh
Lefevre G, Curtis WC (1912) Studies on the reproduction and artificial propagation of freshwater mussels. Bull US Bur Fish 30:105–201
Levin DA (1985) Reproductive character displacement in Phlox. Evol Int J Org Evol 39:1275–1281
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Little TJ, Watt K, Ebert D (2006) Parasite-host specificity: Experimental studies on the basis of parasite adaptation. Evol Int J Org Evol 60:31–38
Lively CM, Craddock C, Vrijenhoek RC (1990) Red queen hypothesis supported by parasitism in sexual and clonal fish. Nature 344:864–866
Mantel N (1967) Detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Maruyama T, Fuerst PA (1984) Population bottlenecks and nonequilibrium models in population genetics. I. Allele numbers when populations evolve from zero variability. Genetics 108:745–763
McCoy KD, Boulinier T, Tirard C, Michalakis Y (2003) Host-dependent genetic structure of parasite populations: differential dispersal of seabird tick host races. Evol Int J Org Evol 57:288–296
Meirmans PG, Hedrick PW (2011) Assessing population structure: F(ST) and related measures. Mol Ecol Resour 11:5–18
Morran LT, Schmidt OG, Gelarden IA, Parrish RC II, Lively CM (2011) Running with the red queen: Host-parasite coevolution selects for biparental sex. Science 333:216–218
Nason JD, Heard SB, Williams FR (2002) Host-associated genetic differentiation in the goldenrod elliptical-gall moth, Gnorimoschema gallaesolidaginis (Lepidoptera: Gelechiidae). Evolution 56:1475–1488
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evol Int J Org Evol 29:1–10
Nieberding C, Morand S, Libois R, Michaux JR (2004) A parasite reveals cryptic phylogeographic history of its host. Proc R Soc Lond B Biol Sci 271:2559–2568
O’Bryhim J, Chong JP, Lance SL, Jones KL, Roe KJ (2012) Development and characterization of sixteen microsatellite markers for the federally endangered species: Leptodea leptodon (Bivalvia: Unionidae) using paired-end Illumina shotgun sequencing. Conserv Genet Resour 4:787–789
Oesch RD (1995) Missouri naiades: a guide to the mussels of Missouri. Missouri Department of Conservation, Jefferson City
Paetkau D, Slade R, Burden M, Estoup A (2004) Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol 13:55–65
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel: population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 65: genetic analysis in excel population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS2: A software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) GENEPOP (version 12): Population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Roe KJ, Boyer SL (2015) A comparison of genetic diversity between sympatric populations of the endangered Winged-Mapleleaf (Quadrula fragosa) and the Pimpleback (Amphinaias pustulosa) in the St. Croix River, USA. Am Malacol Bull 33:52–60
Roe KJ, Simons AM, Hartfield P (1997) Identification of a fish host of the inflated heelsplitter Potamilus inflatus (Bivalvia: Unionidae) with a description of its glochidium. Am Midl Nat 138:48–54
Schwalb AN, Poos MS, Ackerman JD (2011) Movement of logperch-the obligate host fish for endangered snuffbox mussels: implications for mussel dispersal. Aquat Sci 73:223–231
Serb JM (2006) Discovery of genetically distinct sympatric lineages in the freshwater mussel Cyprogenia aberti (Bivalvia: Unionidae). J Molluscan Stud 72:425–434
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Song CB, Near TJ, Page LM (1998) Phylogenetic relations among percid fishes as inferred from mitochondrial cytochrome b DNA sequence data. Mol Phylogenet Evol 10:343–353
Szymanski J (1998) Leptodea leptodon (scaleshell mussel) rangewide status assessment. US Fish and Wildlife Service, Fort Snelling
Tedla S, Fernando CH (1969) Observations on the glochidia of Lampsilis radiata (Gmelin) infesting yellow perch, Perca flavescens (Mitchill) in the Bay of Quinte, Lake Ontario. Can J Zool 47:705–712
Thompson JN (1994) The coevolutionary process. University of Chicago Press, Chicago
Thompson JN (1999) Coevolution and escalation: are ongoing coevolutionary meanderings important? Am Nat 153:S92-S93
USFWS (2010) Scaleshell mussel recovery plan (Leptodea leptodon). US Fish and Wildlife Service, Fort Snelling
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Watters GT (1992) Unionids, fishes, and the species-area curve. J Biogeogr 19:481–490
Zanatta DT, Wilson CC (2011) Testing congruency of geographic and genetic population structure for a freshwater mussel (Bivalvia: Unionoida) and its host fish. Biol J Linn Soc Lond 102:669–685
Zar JH (1984) Biostatistical analysis. Prentice Hall, Englewood Cliffs
Acknowledgements
Funding for this project was provided by the Missouri Department of Conservation and AMEREN UE. Field assistance was provided by the Missouri Department of Conservation and the U.S. Fish and Wildlife Service. In particular we would like to acknowledge the assistance of Steven McMurray, Andy Roberts, Bryan Simmons, Scott Faiman, Josh Hundley, Phillip Pitts, Tim Grace, Nick Girondo, Jennifer Girondo, Ron Burke, Kevin Meneau, Josh Ward, and Rachel Haller for sample collection. We extend our gratitude to John Nason for his advice about the data analysis. We would also like to thank Morgan Sexton, Isaac Jepsen, and Sarah Studeman, undergraduates at Iowa State University, for their laboratory assistance in generating molecular data. Finally, we thank Mary Ashley, Jeanne Serb, and several anonymous reviewers for their valued input on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chong, J.P., Roe, K.J. A comparison of genetic diversity and population structure of the endangered scaleshell mussel (Leptodea leptodon), the fragile papershell (Leptodea fragilis) and their host-fish the freshwater drum (Aplodinotus grunniens). Conserv Genet 19, 425–437 (2018). https://doi.org/10.1007/s10592-017-1015-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-017-1015-x