Abstract
Island populations and populations established by reintroductions are prone to extinction, in part because they are vulnerable to deterministic and stochastic phenomena associated with geographic isolation and small population size. As population size declines, reduced genetic diversity can result in decreased fitness and reduced adaptive potential, which may hinder short- or long-term population viability. We used 32 microsatellite markers to investigate the conservation genetics of a newly established population of Evermann’s Rock Ptarmigan (Lagopus muta evermanni) at Agattu Island, in the western Aleutian Archipelago, Alaska. We found low genetic diversity (observed heterozygosity = 0.41, allelic richness = 2.2) and a small effective population size (N e = 28.6), but a relatively large N e /N ratio = 0.55, which was attributed to multiple paternity in 80% of the broods and low reproductive skew among males (λ = 0.29). Moreover, successful breeding pairs were less related to each other than random male–female pairs. For conservation efforts based on reintroductions, a mating system with high rates of multiple paternity may facilitate retention of genetic diversity, thereby reducing the potential for inbreeding in small or isolated populations. Our results underscore the importance of quantifying genetic diversity and understanding the breeding behavior of translocated populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A common goal for species conservation is maintenance of genetic diversity, but some conservation practices, such as reintroductions, impose both genetic and demographic bottlenecks on newly established populations (Jamieson et al. 2007). Reintroductions are based on translocating organisms into restored areas to re-establish populations in formerly occupied portions of a species’ range (Griffith et al. 1989). Success of reintroduction projects is often measured in terms of demographic performance after establishment (Kaler et al. 2010; Moseby and O’Donnell 2003). Measuring performance based on demographic parameters assesses population dynamics over short time periods, but may overlook the evolutionary consequences of reintroductions on newly established populations.
Small population size imposes genetic bottlenecks on founding populations which can lead to reduced genetic diversity, especially if populations are slow to grow or are established in isolated regions (Keller and Waller 2002). The degree to which founders are related can impact genetic diversity, fitness, evolutionary potential of offspring, and thus the long-term viability of newly established populations (Hedrick et al. 1996; Thuman and Grifith 2005). Mating system and its influence on the effective population size can further impact the rate of loss of genetic variation in small populations.
Loss of genetic variability can be accelerated by social mating systems such as promiscuous lekking where strong reproductive skew among males leads to reduced effective population size (Nooker and Sandercock 2008; Parker and White 1997). However, some mating systems where females mate with multiple males, promote the maintenance of genetic diversity. For example, in cooperative breeding systems, both males and females exhibit strong natal philopatry. To ensure that at least some portion of the offspring are not the result of kin mating (Greenwood 1980), females often mate with multiple males to reduce inbreeding without compromising immediate reproductive opportunities (Brooker et al. 1990). Thus, understanding the mating system of a species under consideration for reintroduction can help managers anticipate genetic outcomes in the population.
Gallinaceous birds are particularly prone to cascading negative effects in small, isolated populations (Briskie and MacIntosh 2004). Reductions in genetic diversity have led to lower egg viability and fecundity rates in isolated populations of Greater Prairie-Chickens (Tympanuchus cupido) and Gunnison Sage-Grouse (Centrocercus minimus) (Stiver et al. 2008; Westemeier et al. 1998, but see Bellinger et al. 2003). Loss of genetic diversity has also led to increased susceptibility to disease and reduced survival of the critically imperiled Attwater’s Prairie-Chicken (T. c. attwateri; Barbosa et al. 2007), and has been implicated in the extinction of the Heath Hen (T. c. cupido; Johnson and Dunn 2006). Therefore, assessment of genetic variability within newly established populations is essential in determining short- and long-term success of translocation projects (Scott and Carpenter 1987).
Rock Ptarmigan (Lagopus muta) are an arctic and alpine species of grouse with a holarctic distribution. The mating system is primarily socially monogamous, with some facultative polygyny (Holder and Montgomerie 1993). Evermann’s Rock Ptarmigan (L. m. evermanni) are an island endemic subspecies restricted to the Near Islands group of the western Aleutian Archipelago, Alaska. By 1936, ptarmigan were extirpated from most of the Near Islands due to the introduction of the exotic arctic fox (Alopex lagopus; Bailey 1993), except for a remnant population at Attu Island (Holder et al. 1999). In 1949, the US Fish and Wildlife Service initiated predator removal programs in the Aleutians. Removal of foxes from Agattu and Attu Islands was completed in 1979 and 1999 (Ebbert and Byrd 2002), but natural dispersal of ptarmigan across the 30 km strait from Attu to Agattu Island did not occur. Consequently, a 4-year reintroduction program was initiated to reestablish a second population of Evermann’s Rock Ptarmigan at Agattu Island by translocating birds from Attu Island.
In this study, we evaluated the genetic diversity, structure, and mating system of the newly established population of Evermann’s Rock Ptarmigan at Agattu Island. Reductions in genetic diversity and fitness are expected after a bottleneck (Jamieson et al. 2007), and one goal of our study was to collect baseline information on genetic diversity, population structure, genetic composition, and effective population size. A second goal was to determine the potential of the genetic mating system to accelerate loss or facilitate retention of genetic diversity among reintroduced birds, given the small population size and geographic isolation that characterize most island populations in the Aleutian Archipelago. For example, female preferences for a subset of males might reduce diversity, whereas high rates of multiple mating could increase female fitness in small populations via reduced risk of inbreeding (Jennions and Petrie 2000). A better understanding of the genetic diversity and mating system in a reintroduced population will aid conservation decisions for ptarmigan in the Aleutian Islands and other endemic populations of birds.
Methods
Study area
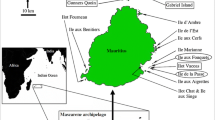
Attu Island (52.85°N, 173.19°E) and Agattu Island (52.43°N, 173.60°E) are part of the Near Island group in the western Aleutian Archipelago in the Alaska Maritime National Wildlife Refuge. Attu Island has an area of 893 km2, and is composed of rugged mountains with a maximum elevation of 861 m. Agattu Island has an area of 225 km2 and is mostly maritime tundra with one mountain range covering approximately one-third of the island with a maximum elevation of 634 m.
Field methods
Five translocations were completed between 2003 and 2006 to reintroduce Evermann’s Rock Ptarmigan to Agattu Island. Ptarmigan were live captured from the Massacre Bay region of Attu Island and translocated to Agattu Island. From May to August of 2005 and 2006, a 2-year post-release demography study was conducted at Agattu Island (Kaler et al. 2010). Despite efforts to translocate mated pairs, in some instances it was not possible to capture both the adult female and male. Ptarmigan are sexually mature at 1 year of age. Upon capture, 40 μl of blood was collected from each bird and placed into 1.5 ml microcentrifuge tubes containing 1 ml of lysis buffer (Seutin et al. 1991). Collection of blood began in 2005 and 2006, 2 years after the initiation of the project; no blood samples were collected in 2003 or 2004. Sampled birds included all ptarmigan translocated from Attu Island to Agattu Island and resident ptarmigan at Agattu Island in 2005 and 2006. Resident Agattu birds included individuals established from translocations in 2003 to 2004 or were the offspring of those translocated birds.
Laboratory methods
We extracted genomic DNA from blood samples using Qiagen DNEasy blood extraction kits following manufacturer protocols (Qiagen USA, Valencia, CA, USA). We selected 32 microsatellites developed for grouse congeners, some of which have been previously cross-amplified in ptarmigan (Lagopus spp.; Table 1). DNA was amplified via the polymerase chain reaction (PCR) in 10 μl volume containing 40 ng of DNA, 2.5 mM MgCl, 0.2 mM dNTP’s, 0.12 µg/µl BSA, 0.8 M Betaine, 0.5 µM forward and reverse primers, 0.2 µM M-13 universal primer, 0.5 units of Promega Flexi Gotaq DNA polymerase, and 1× Promega Flexi Buffer. We used an Applied Biosystems Model 3730 automated sequencer to visualize DNA fragments with fluorescently labeled tags (Operon Biotechnologies, Huntsville, AL) on M-13 universal primers as described by Schuelke (2000). PCR was performed using an Eppendorf Mastercycler Epgradient Thermocycler following published protocols; after amplification, microsatellites were scored using Genemarker 1.6 software. PCR product was multiplexed with up to four microsatellite marker products per tube with the expected size of products all differing by >60 base pairs. We re-analyzed all homozygotes and 20% of heterozygotes to determine if genotyping errors were observer or biochemically based. The presence of null alleles can confound estimates of genetic diversity and parentage assessments (Chakraborty et al. 1992), but we tested all polymorphic loci for the presence of null alleles using Micro-Checker (Oosterhout et al. 2004). Molecular sexing of young was conducted by visualizing introns of the CHD-gene via electrophoresis on 3% high resolution agarose gels after amplification with the sexing primers 2550F and 2718R (Fridolfsson and Ellegren 1999).
Population genetic characteristics
Due to the remote and perilous nature of the field work, samples from the source island, Attu, were collected only from individuals that were translocated. Thus, a direct comparison of genetic diversity of source and reintroduced populations was not possible. Instead, we compared genetic diversity of reintroduced island ptarmigan to mainland ptarmigan to assess variation in genetic diversity regionally. We screened samples with microsatellite loci previously used in this species to compare genetic diversity among birds at Agattu Island and mainland populations of Rock Ptarmigan. Genetic diversity, Hardy–Weinberg Equilibrium (HWE), and linkage disequilibrium were assessed using Genepop 3.4 (Raymond and Rousset 1995), but HWE is not necessarily expected for neutral loci in a translocated population. To avoid problems of resampling among parent and offspring genotypes, independent assessments of genetic diversity were calculated for adults and juveniles. Genetic distance, probability of identity, and probability of exclusion were calculated using GenAlEx6 (Peakall and Smouse 2006). GENETIX 4.05 was used to test if observed estimates of inbreeding (F IS) were significantly different from zero and if the F1 generation had a significantly higher inbreeding coefficient than did the parental generation which could be indicative of a population bottleneck (Belkhir et al. 2004). We used Program Bottleneck (Cornuet and Luikart 1996) to test for excess heterozygosity and a right mode shift in allele frequency, which are indicative of a recent population bottleneck.
We expected that Agattu Island would support a single admixed ptarmigan population. To test this assumption, we explored cryptic population structure using a Bayesian clustering method (Structure 2.2; Pritchard et al. 2000). Inclusion of family groups can influence results for population structure, so only adult birds were used in this analysis (Anderson and Dunham 2008). We tested for structure using all possible combinations of admixture versus no admixture, and correlated versus uncorrelated allele frequency for K = 1–5 populations. Effective population size (N e ) was estimated for the adult population using Program LDNE (Waples 2007).
Breeding ecology
Paternity analyses were performed assuming known maternity using Program Cervus 3.0.3 (Marshall 1998). Prior to paternity assessment, we examined maternity by comparing multi-locus genotypes of chicks to the genotype of the putative mother. A putative mother was defined as the adult female captured with the brood. Each female had one brood in the analysis, and only two chicks had mismatching loci at 1 or 2 of 14 loci. Intraspecific nest parasitism has been reported in ptarmigan but is a rare event (0–6% of nests, Freeland et al. 1995; Martin 1984). Creching and brood mixing are rare among ptarmigan, and only observed late in the breeding season in populations with high breeding densities (Montgomerie et al. 2001; Schmidt 1988). Parasitism and brood mixing were unlikely at Agattu Island because breeding densities were low and chicks were captured near nests within 48 h of hatching (Kaler et al. 2010). Thus, mismatches between mothers and young were best explained as genotyping errors (Morrissey and Wilson 2005).
For paternity analyses, we first conducted an exclusionary analysis based on genotypic mismatching to exclude males as fathers if >1 locus mismatched the chick under analysis. An inclusionary paternity analysis was then conducted for putative fathers not excluded by the exclusionary analysis, using Cervus 3.0.3 (Marshall 1998). Inclusionary methods assign a log-likelihood for paternity based on the highest log-likelihood ratio (ΔLOD, the likelihood that the candidate parent is the true parent, divided by the likelihood that the candidate parent is not the parent), which takes into consideration the background genotypic frequencies within the population and the multilocus genotype of the putative mother. A 90% confidence interval for paternity assignment was generated from population allele frequencies via bootstrapping procedures. Males exceeding the 90% confidence criteria for paternity were assigned as potential fathers to matching offspring (Marshall et al. 1998). Male philopatry or a high degree of relatedness among potential fathers can confound paternity assessments, thereby reducing statistical confidence in assignment for a particular set of genetic markers (Double et al. 1997). Thus, we tested for kin relationships among all sampled males, all males identified as potential fathers, and all males and their assigned offspring using Program Kinship (Goodnight and Queller 1999).
Using the results of paternity assignments, we calculated male reproductive skew based on the λ-index of Kokko and Lindstrom (1997), using Skew Calculator 2003 MAC (Nonacs 2003). The λ-index ranges from equal opportunities for mating among males (λ = 0) to one male obtaining all matings (λ = 1). We grouped males into four non-exclusive categories of reproductive status: males that accounted for the majority of paternity in a brood were ‘primary males’, males with a subset of paternity in the brood were ‘secondary males’, primary and secondary males were collectively termed ‘fathers’, and males without paternity were ‘unmated males’. To test the effects of genetic diversity on male reproductive status between primary, secondary, and unmated males, we used a 1-way ANOVA with orthogonal contrasts in SAS 9.1 (SAS Institute Inc. Cary, NC, USA). Tests of statistical differences between observed group values were also implemented in SAS 9.1, and all descriptive statistics are presented as the mean ± 1 SE.
Results
A total of 75 Evermann’s Rock Ptarmigan were translocated from Attu Island to Agattu Island. Post-release population surveys estimated the ptarmigan population at Agattu Island as ~26 breeding pairs (Kaler et al. 2010). We collected data from 41 adult birds (15 males: 26 females) including 10 females that successfully hatched broods; these data comprised 79% of the estimated breeding population. For 74 chicks captured in the 10 nests at hatching, the observed male: female sex ratio was unexpectedly female-biased at 1:2.04 (binomial test, P = 0.006).
Microsatellite markers
Of 32 microsatellites screened for population genetic analysis, five loci failed to amplify, and 13 loci amplified but were monomorphic in Evermann’s Rock Ptarmigan, despite being polymorphic in other ptarmigan populations (Table 1). Of 14 markers that amplified and were polymorphic, two were linked with other markers and were excluded from estimates of genetic diversity (Table 1). A total of 12 polymorphic microsatellites were suitable for population-wide estimates of genetic diversity. Tests for null alleles indicated weak evidence for null alleles at locus BG16 among adults but not offspring (r adults = 0.044, r offspring = −0.023, r pooled = 0.017). Weak evidence for null alleles was unlikely to impact genetic diversity estimates or parentage assignments, and all 12 polymorphic loci were retained for parentage assignments. Analysis of genetic structure yielded greatest support for a single admixed population at Agattu with correlated allele frequencies (LNP(D) = −15,390.8).
Genetic diversity and relatedness
Population genetic diversity was estimated for adults and young (F1) separately using 12 microsatellite markers (Table 1). Adults had low genetic diversity (HO = 0.41; 95% CI = 0.37–0.46, AR = 2.2; 95% CI = 2.06–2.40), and an inbreeding coefficient significantly less than zero (F IS = −0.28; 95% CI = −0.52 to −0.11, N = 41). The F1 generation had similar levels of diversity (HO = 0.43; 95% CI = 0.37–0.46, AR = 2.2; 95% CI = 1.6–2.8) and a level of inbreeding that did not differ significantly from zero (F IS = 0.07; 95% CI = −0.06–0.11, N = 73). Our estimate of effective population size indicated a relatively small effective population size (N e = 28.6 ± 6.5) but a high N e /N ratio (28.6/52 or 0.55). Bottleneck tests using Wilcoxon tests for excess heterozygosity indicated a significant excess heterozygosity among both the adults (P < 0.001) and chicks (P < 0.001), but no mode shift in allele frequencies. Population-wide estimates of relatedness among all males and females indicated that the population was unrelated overall (r = –0.06 ± 0.02, N = 41). Analysis of relatedness among males indicated low levels of relatedness (r = 0.045 ± 0.034, N = 15); but three males were significantly outbred compared to all other males (r = −0.204, −0.213 and –0.364). Censoring these three individuals and recalculating population-wide relatedness increased the average relatedness of the population (r = 0.157 ± 0.02, N = 38).
Paternity analysis
Paternity analyses were conducted using 14 polymorphic loci, including two linked loci, and allowing for one mismatched locus between father and offspring (Table 1). The 14 loci accounted for 45 alleles yielding a probability of identity per individual of PI = 4.18 × 10−7 and a probability of identity between siblings of PIsibs = 1.2 × 10−3. Our initial paternity analysis included 74 chicks from 10 females and 15 putative fathers. Six of 15 males were excluded as candidate fathers due to incompatible multilocus genotypes. Of the remaining males, the probability that a male with a genotype which was compatible with a chick, but was not the father was low (P = 0.0011).
We accepted paternity assignments at 90% confidence, but more than half were assigned at ≥95% confidence (35 of 63, 56%). The average ΔLOD across all assigned trios exceeded the critical ΔLOD (critical ΔLOD = 0.30; average ΔLOD = 5.63). We were unable to resolve paternity for 11 of 74 chicks. Of 63 chicks with resolved paternity, 77% were perfect matches with both parents. Of 14 chicks which had a single mismatched allele, 11 (89%) were assigned to the same father as the other chicks in the brood. In only three instances did the inclusionary paternal assignment of a chick with a single mismatching paternal loci increase the number of males that a female was inferred to have mated with, but inclusion of these three chicks did not increase the rate of multiple paternity among broods.
Mated pairs successfully producing young were significantly less related to each other (r = –0.13 ± 0.02, N = 20) than random pairs of adults (r = 0.12 ± 0.02, N = 20, two sample t test; t = 2.14, P = 0.049). Multiple paternity rates were high; 80% of broods had paternity assigned to ≥2 males (N = 2 broods with 1 male, 6 broods with 2 males, and 2 broods with 3 males). Overall, 23 chicks (35%) were the product of a female mating with a secondary male. Reproductive skew indices indicated low skew among male reproductive success (λ = 0.29 ± 0.045). However, four males mated and produced chicks with 9 of 10 females (90%), and for 7 of 10 broods (70%), a majority of chicks in the brood were assigned to these four males. Overall, these four males accounted for paternity of 41 of 63 (64%) of the chicks in the analysis.
Analysis of the effect of male genetic diversity on reproductive status indicated that male genetic diversity differed significantly by reproductive status (F 3,71 = 3.57, P = 0.018). Genetic diversity of fathers (HO = 0.51 ± 0.04) was not significantly different than unmated males (HO = 0.48 ± 0.02; F 1,13 = 0.10, P = 0.76). Primary males, however, did have significantly greater heterozygosity (HO = 0.62 ± 0.03) than secondary males (HO = 0.45 ± 0.04, F 1,15 = 9.02, P = 0.004). In addition, we found a significant correlation between male genetic diversity and the number of chicks fathered in the population (r = 0.62, P = 0.03, N = 15). Nevertheless, chicks produced by primary males did not have significantly greater genetic diversity (HO = 0.49 ± 0.04) than chicks produced by secondary males (HO = 0.47 ± 0.03, F 1,21 = 0.42, P = 0.14).
Our sample included eight resident females established from translocations in 2003 to 2004, and two females translocated during the years of our study in 2005–2006. We found no difference in the number of fathers for resident (range = 2–3) or translocated females (range = 1–3). Of 11 chicks with unassigned paternity, eight (73%) were produced by newly translocated females and were the only young to have private alleles. Translocated females initiated nests within 2 days of being relocated to Agattu Island and were likely gravid during transportation from Attu to Agattu. Thus, fathers of these chicks are believed to have originated from Attu.
Discussion
Our analysis of Evermann’s Rock Ptarmigan at Agattu Island revealed the lowest level of genetic diversity ever observed for a wild population of grouse, and are even lower than a relict population of Greater Prairie-Chickens with documented inbreeding depression (HO = 0.57, AR = 3.67; Bouzat et al. 1998). The observed heterozygosity of birds at Agattu Island was 30–50% lower than any mainland population of ptarmigan (Table 2), suggesting that island endemism, perhaps in combination with founder effects, has reduced genetic diversity in these populations (Frankham 1997). Reductions in heterozygosity appear to be a general feature of isolated populations of grouse (Bellinger et al. 2003; Larsson et al. 2008; Stiver et al. 2008; Westemeier et al. 1998), and some vertebrate species have displayed negative impacts of inbreeding depression prior to extinction or extirpation (Spielman et al. 2004). Inbreeding depression has been reported from several wild populations of grouse (Westemeier et al. 1998; Oyler-McCance et al. 2005), and may have been a contributing factor in the extinction of at least one island grouse population, the Heath Hen (T. c. cupido; Johnson and Dunn 2006). In Evermann’s Rock Ptarmigan, low genetic diversity was not associated with reductions in egg viability (Kaler et al. 2010), but it is possible that deleterious genes have been purged from the genome if this unique island subspecies has persisted at low population numbers during its evolutionary history (Holder et al. 1999).
We found evidence that the ptarmigan population at Agattu Island is becoming inbred. The adult, or parental generation, was slightly outbred, whereas the chick, or F1 generation was inbred. We likewise detected an excess of heterozygosity among the chick and adult populations at Agattu Island, which is indicative of a recent population bottleneck (Cornuet and Luikart 1996). Interestingly, we did not observe the expected right mode shift in allele frequencies that is expected to accompany excess heterozygosity in a population which has undergone a recent genetic bottleneck. A likely reason for these observations is that, despite low genetic diversity, ptarmigan at Agattu Island were able to maximize retention of genetic diversity via a combination of relatively low skew in male reproductive success, high rates of multiple paternity, and mate selection.
At Agattu Island, over 80% of the broods and 35% of the chicks were the result of multiple mating which was much higher than extra-pair paternity estimates of 33% of broods and 18% chicks for Willow Ptarmigan (Freeland et al. 1995), and 17% of broods and 5% chicks for White-tailed Ptarmigan (Benson 2002). Low skew in male reproductive success and high rates of multiple paternity can reduce the impacts of isolation and endemism by functionally increasing N e (Pearse and Anderson 2009). Moreover, at low population densities, such as those that occur at Agattu Island, females may not be able to afford to be choosy until after they have already mated, hence the relatively low skew value we observed. Although we lack data on mating chronology, our data are consistent with the notion of females mating randomly with the first male they encounter and then re-mating with higher quality males encountered later in the season. While mates are ultimately selected to maximize individual reproductive success, multiple paternity may prevent bottlenecks in isolated island populations by decreasing the erosion of genetic diversity without foregoing mating opportunities (Sherwin and Moritz 2000).
In addition to low reproductive skew, we found evidence for a genetic contribution to mating behavior and reproductive success. Males and females successfully producing chicks were less related to each other than random pairs which did not produce chicks. Moreover, four males in our sampled population mated with 90% of the females and collectively fathered 64% of the chicks. These same four males had higher observed heterozygosity than other males in the population. We also found a positive correlation between individual male genetic diversity and number of chicks sired. We were unable to determine whether the observed patterns were based on a pre-copulatory mechanism such as kin avoidance, overdominance, or disassociative mating, or to post-copulatory events such as cryptic female choice or genetic incompatibility of close relatives. Nonetheless, our parentage analyses suggest that genetic diversity of male ptarmigan may be an important factor in determining mate choice in females. Ultimately, patterns of mate choice in small populations can affect both individual fitness and population structure (Frankham et al. 2002). Kin avoidance and selection for heterozygous mates should retain remaining genetic diversity in small populations longer than random mating and mitigate some of the genetic erosion that inevitably occurs in small populations (Allendorf and Luikart 2007; Nunney 1993).
Other characteristics of the ptarmigan population may enhance the overall genetic diversity and prospects for long term survival at Agattu Island. First, inclusion of three outbred individuals from Attu Island reduced the average relatedness and inbreeding for the newly established population. Second, several chicks had private alleles not found among any of the adults sampled at Agattu Island. These chicks could have been sired by males at Attu Island due to the capture of gravid females, or by males that we were unable to sample at Agattu Island. We believe the first hypothesis is more plausible for two reasons: (1) Mothers of the chicks with private alleles initiated nests 2 days after translocation to Agattu Island. This time period is too short for these chicks to be attributed to Agattu males (Kaler et al. 2010). In ptarmigan, nest initiation is 4–12 days post copulation because of the time required for embryonic follicles to develop into eggs post fertilization (Thomas 1986). (2) Some females laid eggs while in transit from Attu to Agattu Island, indicating that some females were indeed inseminated by Attu males. Ultimately, female mate choice and high rates of multiple paternity contributed to an effective population size greater than expected for a small insular population and thus a large N e /N ratio. Our results are encouraging because genetic diversity is lost more slowly in populations with high N e/N ratios (Nunney 1995). Translocation of outbred males and gravid females were unintended benefits of our field protocols, but combined with mate choice, the net effects on genetic diversity and potential reductions in inbreeding may increase both the short- and long-term population viability of Evermann’s Rock Ptarmigan at Agattu Island.
One unexpected finding in our study was a female-biased sex ratio among chicks. Several lines of evidence suggest that females are able to control the primary sex ratio of offspring in some species of vertebrates (Clutton-Brock and Iason 1986; Komdeur et al. 2002). For example, establishment of a translocated population of Seychelles Warblers (Acrocephalus sechellensis) to an unpopulated island resulted in a female-biased primary sex ratio, and mated pairs switched from male-biased to female-biased clutches after translocation (Komdeur et al. 1997). We lack data on sex-specific survival but if a female-biased sex ratio persists in the adult population, then the potential exists for rapid population growth, which would further buffer the population from loss of genetic diversity (Allendorf and Luikart 2007).
Conservation biology is frequently described as a crisis discipline, which must be holistic and integrate information from different fields of study (Soulé 1985). Nevertheless, the utility of population genetics in species conservation and management has been debated (Caughley 1994; Brookes 1997; Vernesi et al. 2008). Our genetic analysis of a newly established Rock Ptarmigan population at Agattu Island highlights how characteristics of the mating system and population genetics combined with translocation strategies can positively influence management success of conservation efforts. Consequently, the ultimate success of a translocation effort may hinge upon understanding the baseline genetic diversity of source and translocated populations as well as the genetic mating system. In the case of ptarmigan translocations in the Aleutian Islands, fortuitous translocation of genetically outbred and gravid individuals, restricted mating between close relatives, and high rates of multiple paternity should improve the probability that the newly established populations will avoid the deleterious effects of inbreeding depression. In the future, to facilitate successful management, conservation, and establishment of evolutionarily stable populations, translocation and management efforts should devise conservation strategies that include population genetics as part of the core project objectives.
References
Allendorf FW, Luikart G (2007) Conserving global biodiversity conservation and the genetics of populations. Blackwell Publishing, Oxford
Anderson EC, Dunham KK (2008) The influence of family groups on inferences made with Program Structure. Mol Ecol Res 8:1219–1229
Bailey E (1993) Introduction of foxes to Alaskan Islands: history, effects on avifauna, and eradication. U.S. Fish and Wildlife Service Resource Publication 191, Anchorage
Barbosa T, Zavala G, Cheng S, Villegas P (2007) Full genome sequence and some biological properties of reticuloendotheliosis virus strain APC-566 isolated from endangered Attwater’s Prairie-Chicken. Virus Res 124:68–77
Bech N, Boissier J, Drovetski S, Novoa C (2009) Population genetic structure of Rock Ptarmigan in the ‘sky islands’ of French Pyrenees: implications for conservation. Anim Conserv 12:138–146
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier
Bellinger RM, Johnson JA, Toepfer J, Dunn PO (2003) Loss of genetic variation in Greater Prairie-Chickens following a population bottleneck in Wisconsin, U.S.A. Conserv Biol 17:717–724
Benson D (2002) Low extra-pair paternity in White-tailed Ptarmigan. Condor 104:192–197
Bouzat JL, Cheng HH, Lewins HA, Westemeier RL, Brawn JD, Paige KN (1998) Genetic evaluation of a demographic bottleneck in the Greater Prairie-Chicken. Conserv Biol 12:836–843
Briskie JV, MacIntosh M (2004) Hatching failure increases with severity of bottlenecks in birds. Proc Natl Acad Sci USA 101:558–561
Brooker MG, Rowley I, Adams M, Baverstock PR (1990) Promiscuity: an inbreeding avoidance mechanism in a socially monogamous species? Behav Ecol Sociobiol 26:191–199
Brookes M (1997) A clean break. New Sci 156:64
Caizergues A, Dubois S, Mondor G, Loiseau A, Ellison LN, Raspluss J-Y (2001) Isolation and characterization of microsatellite loci in Black Grouse (Tetrao tetrix). Mol Ecol Notes 1:36–38
Caizergues A, Bernard-Laurent A, Brenot JF, Ellison LN, Raspluss J-Y (2003) Population genetic structure of Rock Ptarmigan Lagopus muta in Northern and Western Europe. Mol Ecol 12:2267–2274
Caughley G (1994) Directions in conservation biology. J Anim Ecol 63:215–244
Chakraborty R, Deandrade M, Daiger SP, Budowle B (1992) Apparent heterozygote deficiencies observed in DNA typing data and their implications in forensic applications. Ann Hum Genet 56:45–57
Cheng HH, Levin I, Vallejo RL, Khatib H, Dodgson JB, Crittenden LB, Hillel J (1995) Development of a genetic map of the chicken with markers of high utility. Poult Sci 74:1855–1874
Clutton-Brock TH, Iason GR (1986) Sex ratio variation in mammals. Quant Rev Biol 61:339–374
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Double MC, Cockburn A, Smouse PE (1997) Exclusion probabilities for single locus paternity analysis when related males compete for matings. Mol Ecol 6:1155–1166
Ebbert SE, Byrd GV (2002) Eradications of invasive species to restore natural biological diversity on Alaska Maritime National Wildlife Refuge. In: Veitch CR, Clout MN (eds) Turning the tide: the eradication of invasive species. IUCN SSC invasive species specialist group, IUCN, Gland
Frankham R (1997) Do island populations have less genetic variation than mainland populations? Heredity 78:311–327
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, New York
Freeland JR, Hannon SJ, Dobush G, Boag PT (1995) Extra-pair paternity in Willow Ptarmigan broods: measuring costs of polygyny to males. Behav Ecol Sociobiol 36:349–355
Fridolfsson A, Ellegren H (1999) A simple and universal method for molecular sexing of non-ratite birds. J Avian Biol 30:116–121
Goodnight KF, Queller DC (1999) Computer software for performing likelihood tests of genetic pedigree relationships using genetic markers. Mol Ecol 8:1231–1234
Greenwood PJ (1980) Mating systems, philopatry, and dispersal in birds and mammals. Anim Behav 28:1140–1162
Griffith BJ, Scott M, Carpenter JW, Reed C (1989) Translocation as a species conservation tool: status and strategy. Science 245:477–480
Hedrick PW, Lacey RC, Allendorf FW, Soulé ME (1996) Directions in conservation biology: comments on Caughley. Conserv Biol 10:1312–1320
Holder K, Montgomerie R (1993) Rock Ptarmigan (Lagopus muta). In: Poole A, Gill F (eds) The Birds of North America, No. 51. The Academy of Natural Sciences, Washington
Holder K, Montgomerie R, Friesen VL (1999) A test of the glacial refugium hypothesis using patterns of mitochondrial and nuclear DNA sequence variation in Rock Ptarmigan (Lagopus muta). Evolution 53:1936–1950
Jamieson IG, Tracy LN, Fletcher D, Armstrong DP (2007) Moderate inbreeding depression in a reintroduced population of North Island robins. Anim Conserv 10:95–102
Jennions MD, Petrie M (2000) Why do females mate multiply: a review of the genetic benefits. Biol Rev 75:21–64
Johnson J, Dunn PO (2006) Low genetic variation in the Heath Hen prior to extinction and implications for the conservation of prairie chicken populations. Conserv Genet 7:37–48
Kaler RSA, Ebbert SE, Braun CE, Sandercock BK (2010) Demography of a reintroduced population of Evermann’s Rock Ptarmigan in the Aleutian Islands. Wilson J Ornith 122:1–14
Keller L, Waller D (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Kokko WD, Lindstrom J (1997) Measuring mating skew. Am Nat 149:794–799
Komdeur J, Daan S, Tinbergen J, Mateman AC (1997) Extreme adaptive modification in sex ratio of the Seychelles Warbler’s eggs. Nature 385:522–525
Komdeur J, Magrath MJL, Krackow S (2002) Pre-ovulation control of hatchling sex ratio in the Seychelles warbler. Proc Roy Soc Lon B 269:1067–1072
Larsson JK, Jansman HAH, Segelbacher G, Höglund J, Koelewijn HP (2008) Genetic impoverishment of the last Black Grouse (Tetrao tetrix) population in the Netherlands: detectable only with a reference from the past. Mol Ecol 17:1897–1904
Marshall T (1998) Program Cervus (ver 3.0.3). Field Genetics Ltd. www.fieldgenetics.com
Marshall T, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
Martin K (1984) Intraspecific nest parasitism in Willow Ptarmigan. J Field Ornithol 55:250–251
Montgomerie R, Lyon BE, Holder K (2001) Dirty ptarmigan: behavioral modification of sexually selected trait. Behav Ecol 12:429–438
Morrissey MB, Wilson AJ (2005) The potential costs of accounting for genotypic errors in molecular parentage analyses. Mol Ecol 14:4111–4121
Moseby KE, O’Donnell E (2003) Reintroduction of the greater bilby, Macrotis lagotis (Reid) (Marsupialia: Thylacomidae), to northern and South Australia: survival, ecology and notes on reintroduction protocol. Wildl Resour 30:15–27
Nonacs P (2003) Measuring the reliability of skew indices: is there one best index? Anim Behav 65:615–627
Nooker JK, Sandercock BK (2008) Phenotypic correlates and survival consequences of male mating success in lek-mating Greater Prairie-Chickens (Tympanuchus cupido). Behav Ecol Sociobiol 62:1377–1388
Nunney L (1993) The influence of mating system and overlapping generations on effective population size. Evolution 47:1329–1341
Nunney L (1995) Measuring the ratio of effective population size to adult numbers using genetic and ecological data. Evolution 49:389–392
Oosterhout CV, Hutchinson WF, Wills DP, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Oyler-McCance SJ, John JS, Taylor SE, Apa AD, Quinn TW (2005) Population genetics of Gunnison Sage-Grouse: implications for management. J Wildl Manag 69:630–637
Parker PG, White TA (1997) Mating systems, effective population size, and conservation in natural populations. In: Clemmons JR, Buchholz R (eds) Behavioral approaches to conservation in the wild. Cambridge University Press, Cambridge, pp 243–261
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pearse DE, Anderson EC (2009) Multiple paternity increases effective population size. Mol Ecol 18:3124–3127
Piertney SB, Dallas JF (1997) Isolation and characterization of hyper-variable microsatellites in Red Grouse Lagopus lagopus scoticus. Mol Ecol 6:93–95
Piertney SB, Höglund J (2001) Polymorphic microsatellite DNA markers in Black Grouse (Tetrao tetrix). Mol Ecol Notes 1:303–304
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) GENEPOP (version 3.4): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Sahlman T, Segelbacher G, Höglund J (2009) Islands in the ice: colonization routes for rock ptarmigan to the Svalbard archipelago. Ecography 32:840–848
Schmidt RK (1988) Behavior of White-tailed Ptarmigan during the breeding season. In: Bergerud AT, Gatson MW (eds) Adaptive strategies and population ecology of northern grouse, vol 2. University of Minnesota Press, Minneapolis, pp 270–299
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotech 18(2):233–234
Scott JM, Carpenter JW (1987) Release of captive-reared or translocated endangered birds: what do we need to know? Auk 104:544–545
Seutin G, White BN, Boag PT (1991) Preservation of avian blood and tissue samples for DNA analysis. Can J Zool 69:82–90
Sherwin WB, Moritz C (2000) Managing and monitoring genetic erosion. In: Young AG, Clarke GM (eds) Genetics demography, and viability of fragmented populations. Cambridge University Press, Cambridge, pp 9–34
Soulé ME (1985) What is conservation biology? Bioscience 11:727–734
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci USA 101:15261–15264
Stiver JR, Apa AD, Remington TE, Gibson RM (2008) Polygyny and female breeding failure reduce effective population size in the lekking Gunnison Sage-Grouse. Biol Conserv 141:472–481
Taylor SE, Oyler-McCance SJ, Quinn TW (2003) Isolation and characterization of microsatellite loci in Greater Sage-Grouse (Centrocercus urophasianus). Mol Ecol Notes 3:262–264
Thomas VG (1986) Body condition, ovarian hierarchies, and their relation to egg formation in Anseriformes and Galliform species. In: Ouellet H (ed) Acta XIX Congressus Internationalis Ornithologici. University of Ottawa Press, Ottawa, pp 353–363
Thuman KA, Grifith SC (2005) Genetic similarity and the nonrandom distribution of paternity in a genetically highly polyandrous shorebird. Anim Behav 69:765–770
Vernesi C, Bruford MW, Bertorelle G, Pecchioli E, Rizzoli A, Hauffe HC (2008) Where’s the conservation in conservation genetics? Conserv Biol 22:802–804
Waples RS (2007) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Notes 7:167–184
Westemeier R, Braun J, Simpson S, Esker T, Jansen R, Walk J, Kershner E, Bouzat J, Paige K (1998) Tracking the long-term decline and recovery of an isolated population. Science 282:1695–1698
Acknowledgments
Thanks to C.E. Braun, W.P. Taylor, and S.E. Ebbert for initiating the Rock Ptarmigan translocation project and to L.A. Kenney, G.T. Wann and other technicians for field assistance. Captain K.D. Bell and the crew of the Tiglax provided safe transport in the Aleutian Islands and the U.S. Coast Guard LORAN Station provided field housing at Attu Island. Funding was provided by the USFWS Alaska Maritime National Wildlife Refuge, U.S. Missile Defense Agency, and Kansas State University. Capture and handling of birds was approved by the Kansas State University Institutional Animal Care and Use Committee (protocol # 2361) and wildlife permits by the State of Alaska.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Gregory, A.J., Kaler, R.S.A., Prebyl, T.J. et al. Influence of translocation strategy and mating system on the genetic structure of a newly established population of island ptarmigan. Conserv Genet 13, 465–474 (2012). https://doi.org/10.1007/s10592-011-0300-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-011-0300-3