Abstract
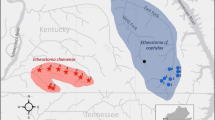
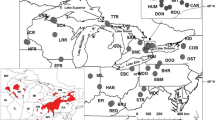
Shortnose sturgeon Acipenser brevirostrum is federally listed as ‘‘an endangered species threatened with extinction’’ in the U.S. but its listing status is currently under review. As part of this process, the U.S. National Marine Fisheries Service will determine if shortnose sturgeon are divided into Distinct Population Segments (DPS) across its distribution. In this regard, we sought to determine if shortnose sturgeon occur in genetically “discrete population segments,” and if so, the boundaries of each. We used mitochondrial DNA (mtDNA) control region sequence analysis to assess the genetic discreteness of 14 of 19 river populations that were recommended as DPS in the 1998 Final Recovery Plan for Shortnose Sturgeon. Nine of the 14 proposed DPS proved significantly discrete (P < 0.05 after Bonferoni correction) from both of their bracketing populations, the exceptions being those in the Penobscot River, Chesapeake Bay, Cooper River, and Ogeechee River (our sample from the Cape Fear River was insufficient to statistically analyze). Haplotype frequencies in the newly “rediscovered” Penobscot River collection were almost identical to those in the proximal Kennebec River system. Genetic data in combination with tagging results suggest that shortnose sturgeon in the Penobscot River are probably migrants from the Kennebec. Likewise, shortnose sturgeon found today within the Chesapeake Bay appear to be migrants from the Delaware River. While haplotype frequencies in the remnant Santee River population in Lake Marion differed significantly from those in nearby Winyah Bay, they did not differ significantly from those in the Cooper River. This suggests that the Cooper River harbors descendants of the Santee River population that are unable to access their historical spawning locales. The Ogeechee River collection was not genetically distinct from that in the nearby Savannah River, suggesting that it may host descendants of hatchery-reared individuals of Savannah River ancestry. Our genetic results indicate that most, but not all, rivers with shortnose sturgeon host genetically discrete populations, constituting important information in the consideration of DPS designations. However, shortnose sturgeon migrations through coastal waters to proximal rivers and release of hatchery-reared fish may confound results from genetic studies such as ours and lead to the possible misidentification of discrete population segments.




Similar content being viewed by others
References
Bain MD (2001) Sturgeon of the Hudson River: ecology of juveniles. Final report to the Hudson River Foundation, New York
Beechie T, Buhle E, Ruckelhaus M, Fullerton A, Holsinger L (2006) Hydrologic regime and the conservation of salmon life history diversity. Biol Conserv 130:560–572. doi:10.1016/j.biocon.2006.01.019
Bohonak AJ (1999) Dispersal, gene flow, and population structure. Q Rev Biol 74:21–45. doi:10.1086/392950
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene geneaologies. Mol Ecol 9:1657–1659. doi:10.1046/j.1365-294x.2000.01020.x
Collins MR, Rogers SG, Smith TIJ, Moser ML (2000) Primary factors affecting sturgeon populations in the southeastern U.S.: fishing mortality and degradation of essential habitat. Bull Mar Sci 66:917–928
Collins MR, Cooke D, Post B, Crane J, Bulak J, Smith TIJ, Greig TW, Quattro JM (2003) Shortnose sturgeon in the Santee-Cooper Reservoir system, South Carolina. Trans Am Fish Soc 132:1244–1250. doi:10.1577/T02-036
Cooke DW, Leach SD (2003) Beneficial effects of increased river flow and upstream fish passage on anadromous alosine stocks. Am Fish Soc Symp 35:331–338
Cooke DW, Leach SD (2004a) Implications of a migration impediment on shortnose sturgeon spawning. N Am J Fish Manag 24:1460–1468. doi:10.1577/M03-141.1
Cooke DW, Leach SD (2004b) Santee Cooper FERC studies updated. Report (submitted to). South Carolina Department of Natural Resources, Bonneau, SC
Cooke DW, Leach SD, Isely JJ (2002) Behavior and lack of upstream passage of shortnose sturgeon at a hydroelectric facility and navigation lock complex. In: Van Winkle W, Anders PJ, Secor DH, Dixon DA (eds) Biology, management, and protection of North American Sturgeon, vol 28. American Fisheries Society Symposium, Oshkosh, WI, pp 101–110
Cooke DW, Kirk JP, Morrow JV Jr, Leach SD (2004) Population dynamics of a migration limited shortnose sturgeon population. In: Proceedings of Annual Conference of Southeastern Association for Fish and Wildlife Agencies, vol 58, pp 82–91
COSEWIC (2005) COSEWIC assessment and update status report on the shortnose sturgeon Acipenser brevirostrum in Canada. Committee on the Status of Endangered Wildlife in Canada, Ottawa, 27 pp
Dadswell MJ (1976) Biology of the shortnose sturgeon (Acipenser brevirostrum) in the Saint John River estuary, New Brunswick, Canada. Transactions of the Atlantic Chapter of Canadian Society of Environmental Biology Annual Meeting 1975, pp 20–72
Dadswell MJ (1979) Biology and population characteristics of the shortnose sturgeon, Acipenser brevirostrum LeSueur 1818 (Osteichthyes: Acipenseridae), in the Saint John River estuary, New Brunswick, Canada. Can J Zool 57:2186–2210. doi:10.1139/z79-287
Dadswell MJ, Taubert BD, Squiers TS, Marchette D, Buckley J (1984) Synopsis of biological data on shortnose sturgeon, Acipenser brevirostrum Le Sueur 1818. FAO Fish Synop 140:1–45
Duncan MS, Isely JJ, Cooke DW (2004) Evaluation of shortnose sturgeon spawning in the Pinopolis Dam tailrace, South Carolina. N Am J Fish Manag 24:932–938. doi:10.1577/M03-131.1
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.1: an integrated software package for population genetic analysis. Evol Bioinform Online 1:47–50
Felsenstein J (1985) Confidence limits on phylogenies: an approach using bootstrap. Evolution Int J Org Evolution 39:783–791. doi:10.2307/2408678
Finney ST, Isely JJ, Cooke DW (2006) Upstream migration of two pre-spawning shortnose sturgeon passed upstream of Pinopolis Dam, Cooper River, South Carolina. Southeast Nat 5:36–375. doi:10.1656/1528-7092(2006)5[369:UMOTPS]2.0.CO;2
Fu Y-X (1997) Statistical tests of neutrality against population growth, hitchhiking and background selection. Genetics 143:557–570
Gibbons JJ, Leach SD, Cooke DW, Wirgin II (2007) Santee-Cooper basin shortnsoe sturgeon monitoring II. Final programmatic report to National Fish and Wildlife Foundation, 49 pp
Grunwald C, Stabile J, Waldman JR, Wirgin II (2002) Population genetics of shortnose sturgeon Acipenser brevirostrum based on mitochondrial DNA control region sequences. Mol Ecol 11:1885–1898. doi:10.1046/j.1365-294X.2002.01575.x
Grunwald C, Maceda L, Waldman J, Stabile J, Wirgin I (2007) Conservation of Atlantic sturgeon Acipenser oxyrinchus oxyrinchus: delineation of stock structure and distinct population segments. Conserv Genet 9:1111–1124. doi:10.1007/s10592-007-9420-1
Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589
Kjerfve B (1975) The Santee-Cooper: a study of estuarine manipulations. In: Wiley M (ed) Estuarine processes. Academic Press, NY, pp 44–56
Kynard B (1997) Life history, latitudinal patterns, and status of shortnose sturgeon, Acipenser brevirostrum. Environ Biol Fish 48:319–334. doi:10.1023/A:1007372913578
Marchette DE, Smiley R (1982) Biology and life history of incidentally captured shortnose sturgeon, Acipenser brevirostrum in South Carolina. Report to South Carolina Wildlife and Marine Resources Research Institute, Charleston, South Carolina, 55 pp
McDowall RM (2001) Anadromy and homing: two life-history traits with adaptive synergies in salmonid fishes? Fish Fish 2:78–85. doi:10.1046/j.1467-2979.2001.00036.x
McElroy D, Moran P, Bermingham E, Kornfield I (1992) REAP: an integrated environment for the manipulation of and phylogenetic analysis of restriction data. J Hered 83:157–158
Miller MP (1997) Tools for population genetic analyses (TFPGA) 1.3; a Windows program for the analysis of allozyme and molecular population genetic data
National Marine Fisheries Service (1987) Status review of shortnose sturgeon (Acipenser brevirostrum LeSueur 1818). Listed under the Endangered Species Act of 1973. 31 pp
National Marine Fisheries Service (1996) Status review of shortnose sturgeon in the Androscoggin and Kennebec River. Northeast Regional Office, National Marine Fisheries Service. 25 pp
National Marine Fisheries Service (1998) Recovery plan for the shortnose sturgeon (Acipenser brevirostum). Prepared by the shortnose sturgeon recovery team for the National Marine Fisheries Service, Silver Spring, MD. 104 pp
Nei M (1972) Genetic distances between populations. Am Nat 106:283–292. doi:10.1086/282771
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, NY
Nielsen R, Wakeley J (2001) Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158:885–896
Parker EL (2007) Ontogeny and life history of shortnose sturgeon (Acipenser brevirostrum Lesueur 1818): effects of latitudinal variation and water temperature. Ph.D. Thesis, University of Massachusetts, Amherst, Massachusetts
Quattro JM, Greig TW, Coykendall DK, Bowen BW, Baldwin JD (2002) Genetic issues in aquatic species management: the shortnose sturgeon (Acipenser brevirostrum) in the southeastern United States. Conserv Genet 3:155–166. doi:10.1023/A:1015275602748
Roff DA, Bentzen P (1989) The statistical analysis of mitochondrial DNA polymorphisms: X 2 and the problem of small samples. Mol Biol Evol 6:539–545
Saghai-Maroof MA, Soliman KM, Jorgneson RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018. doi:10.1073/pnas.81.24.8014
Shepherd TD, Litvak MK (2004) Density-dependent habitat selection and the ideal free distribution in marine fish spatial dynamics: considerations and cautions. Fish Fish 5:141–152. doi:10.1111/j.1467-2979.2004.00143.x
Slatkin M (1985) Gene flow in natural populations. Annu Rev Ecol Syst 16:393–430. doi:10.1146/annurev.ecolsys.16.1.393
Smith TIJ, Collins MC, Post WC, McCord JW (2002a) Stock enhancement of shortnose sturgeon. Am Fish Soc Symp 28:31–44
Smith TIJ, McCord JW, Collins MR, Post WC (2002b) Occurrence of stocked shortnose sturgeon Acipenser brevirostrum in non-target rivers. J Appl Ichthyology 18:470–474. doi:10.1046/j.1439-0426.2002.00412.x
Squiers TS (2003) State of Maine 2003 Atlantic sturgeon compliance report to the Atlantic States Marine Fisheries Commission. Report submitted to the Atlantic States Marine Fisheries Commission, October 31, 2003, Washington, DC
Squiers Jr TS (1993) Assessment of potential shortnose sturgeon spawning sites in the upper tidal reach of the Androscoggin River. Final report of results to the Maine Department of Transportation
Squiers TS, Smith M (1979) Distribution and abundance of shortnose sturgeon in the Kennebec River estuary. Final report to the National Marine Fisheries Service, Gloucester, MA
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tajima F (1993) Measurement of DNA polymorphism. In: Takahata N, Clark GS (eds) Mechanisms of molecular evolution. Introduction to molecular paleopopulation biology. Japan Scientific Societies Press, Sinauer Associates, Sunderland
Thompson GG (1991) Determining minimum viable populations under the Endangered Species Act. National Marine Fisheries Service, Technical Memorandum NMFS F/NWC-198. 78 pp
USACE (U.S. Army Corp of Engineeers) (1975) Final environmental statement, Cooper River Rediversion Project, Charleston Harbor, South Carolina. USACE, Charleston, SC
Waldman JR, Grunwald C, Stabile J, Wirgin I (2002) Impacts of life history and biogeography on genetic stock structure in Atlantic Sturgeon, Acipenser oxyrinchus oxyrinchus, Gulf sturgeon A. oxyrinchus desotoi, and shortnose sturgeon, A. brevirostrum. J Appl Ichthyology 18:509–518. doi:10.1046/j.1439-0426.2002.00361.x
Walsh MG, Bain MB, Squiers T Jr, Waldman JR, Wirgin I (2001) Morphological and genetic variation among shortnose sturgeon Acipenser brevirostrum from adjacent and distant rivers. Estuaries 24:41–48. doi:10.2307/1352811
Welsh SA, Mangold MF, Skjeveland JE, Spells AJ (2002) Distribution and movement of shortnose sturgeon (Acipenser brevirostrum) in the Chesapeake Bay. Estuaries 25:101–104
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution Int J Org Evolution 38:1358–1370. doi:10.2307/2408641
Wippelhauser G (2003) Striped bass and American shad restoration and monitoring. Project #F-41-R-9 annual report
Wirgin I, Grunwald C, Carlson E, Stabile J, Peterson DL, Waldman JR (2005) Range-wide population structure of shortnose sturgeon Acipenser brevirostrum based on sequence analysis of the mitochondrial DNA control region. Estuaries 28:406–421. doi:10.1007/BF02693923
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Ziegeweid JR, Jennings CA, Peterson DL, Black MC (2008) Effects of salinity, temperature, and weight on the survival of young-of-year shortnose sturgeon. Trans Am Fish Soc 137:1490–1499. doi:10.1577/T07-046.1
Acknowledgments
We thank M. Litvak, S. Usvyatsov, M. Kinnison, S. Fernandez, D. Cooke, and J. Gibbons for acquisition of the new samples analyzed in this project. We thank D. Hartley, J. Gibbons, and K. Damon-Randall for commenting on earlier drafts of this manuscript. We acknowledge support of the Protected Species Division of the National Marine Fisheries Service, Julie Carter of the National Ocean Service/National Centers for Coastal Ocean Science, Marine Forensics Archive for providing tissue samples, and the Molecular Facility Core of the New York University National Institute of Environmental Health Sciences Center ES00260 for the use of its shared instrumentation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wirgin, I., Grunwald, C., Stabile, J. et al. Delineation of discrete population segments of shortnose sturgeon Acipenser brevirostrum based on mitochondrial DNA control region sequence analysis. Conserv Genet 11, 689–708 (2010). https://doi.org/10.1007/s10592-009-9840-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-009-9840-1