Abstract
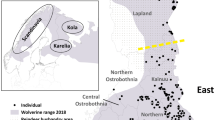
We examined the genetic diversity and structure of wolf populations in northwestern Russia. Populations in Republic of Karelia and Arkhangelsk Oblast were sampled during 1995–2000, and 43 individuals were genotyped with 10 microsatellite markers. Moreover, 118 previously genotyped wolves from the neighbouring Finnish population were used as a reference population. A relatively large amount of genetic variation was found in the Russian populations, and the Karelian wolf population tended to be slightly more polymorphic than the Arkhangelsk population. We found significant inbreeding (F = 0.094) in the Karelian, but not in the Arkhangelsk population. The effective size estimates of the Karelian wolf population based on the approximate Bayesian computation and linkage disequilibrium methods were 39.9 and 46.7 individuals, respectively. AMOVA-analysis and exact test of population differentiation suggested clear differentiation between the Karelian, Arkhangelsk and Finnish wolf populations. Indirect estimates of gene flow based on the level of population differentiation (ϕ ST = 0.152) and frequency of private alleles (0.029) both suggested a low level of gene flow between the populations (Nm = 1.4 and Nm = 3.7, respectively). Assignment analysis of Karelian and Finnish populations suggested an even lower number of recent migrants (less than 0.03) between populations, with a larger amount of migration from Finland to Karelia than vice versa. Our findings emphasise the role of physical obstacles and territorial behaviour in creating barriers to gene flow between populations in relatively limited geographical areas, even in large-bodied mammalian species with long-distance dispersal capabilities and an apparently continuous population structure.






Similar content being viewed by others
References
Anonymous (2005) Management plan for the wolf population in Finland. Ministry of Agriculture and Forestry Publications 11b/2005. Available at: http://wwwb.mmm.fi/julkaisut/julkaisusarja/2005/MMMjulkaisu2005_11b.pdf. Accessed 14 July 2008
Aspi J, Roininen E, Ruokonen M, Kojola I, Vilà C (2006) Genetic diversity, population structure, effective population size, and demographic history of the Finnish wolf population. Mol Ecol 15:1561–1576. doi:10.1111/j.1365-294X.2006.02877.x
Beaumont MA, Zhang W, Balding DJ (2002) Approximate Bayesian computation in population genetics. Genetics 162:2025–2035
Belkhir K, Borsa P, Chikhi L, Raufaste N, Catch F (2004) Genetix 4.0.5.2., Software under Windows™ for the genetics of the populations. Laboratory genome, populations, interactions, CNRS UMR 5000. University of Montpellier II, Montpellier, France
Bensch S, Andrén H, Hansson B, Pedersen HC, Sand H, Sejberg D et al (2007) Selection for heterozygosity gives hope to a wild population of inbred wolves. PLoS ONE 1:e72. doi:10.1371/journal.pone.0000072
Berry O, Tocher MD, Sarre SD (2004) Can assignment tests measure dispersal? Mol Ecol 13:551–561. doi:10.1046/j.1365-294X.2004.2081.x
Black WC, Kraftsur ES (1985) A FORTRAN programme for the calculation and analysis of two-locus linkage disequilibrium coefficients. Theor Appl Genet 70:491–496. doi:10.1007/BF00305981
Boitani L (2003) Wolf conservation and recovery. In: Mech LD, Boitani L (eds) Wolves. Behavior, ecology and conservation. University of Chicago Press, Chicago, pp 317–340
Carmichael LE, Nagy JA, Larter NC, Strobeck C (2001) Prey specialization may influence patterns of gene flow in wolves of the Canadian Northwest. Mol Ecol 10:2787–2798
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Danilov PI (2005) Game animals of Karelia. Ecology, resources, management, protection. Nauka, Moscow 338 pp (in Russian)
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure from multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Francisco LV, Langston AA, Mellersh CS, Neal CL, Ostrander EA (1996) A class of highly polymorphic tetranucleotide repeats for canine genetic mapping. Mamm Genome 7:359–362. doi:10.1007/s003359900104
Fredholm M, Winterø AK (1995) Variation of short tandem repeats within and between species belonging to the Canidae family. Mamm Genome 6:11–18. doi:10.1007/BF00350887
Garza C, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318. doi:10.1046/j.1365-294x.2001.01190.x
Geffen E, Anderson MJ, Wayne RK (2004) Climate and habitat barriers to dispersal in the highly mobile grey wolf. Mol Ecol 13:2481–2491. doi:10.1111/j.1365-294X.2004.02244.x
Goudet J (2001) FSTAT: a program to estimate and test gene diversities and fixation indices (version 2.9.3)
Harley E (2004) AGARst: a program for calculating Allele frequencies, Gst and Rst from microsatellite data and outputting files formatted for other programs such as ‘genepop’ and ‘rstcalc’. http://web.uct.ac.za/depts/chempath/genetic.htm. Accessed 6 Dec 2006
Kojola I, Määttä E (2004) Suurpetojen lukumäärä ja lisääntyminen vuonna 2003 (The number and reproduction of large carnivores in Finland in 2003). Riistantutkimuksen Tiedote 194:1–7 (in Finnish)
Kojola I, Aspi J, Hakala A, Heikkinen S, Ilmoni C, Ronkainen S (2006) Dispersal in an expanding wolf population. J Mammal 87:281–286. doi:10.1644/05-MAMM-A-061R2.1
Laikre L (1999) Conservation genetics of Nordic carnivores: lessons from zoos. Hereditas 130:203–216. doi:10.1111/j.1601-5223.1999.00203.x
Liberg O (2005) Genetic aspects of viability in small wolf populations with special emphasis on the Scandinavian wolf population. Report no 5436. The Swedish Environmental Protection Agency
Liberg O, Andre’n H, Pedersen HC et al (2005) Severe inbreeding depression in a wild wolf Canis lupus population. Biol Lett 1:17–20. doi:10.1098/rsbl.2004.0266
Lucchini V, Galov A, Randi E (2004) Evidence of genetic distinction and long-term population decline in wolves (Canis lupus) in the Italian Apennines. Mol Ecol 13:523–536. doi:10.1046/j.1365-294X.2004.02077.x
Manel S, Gaggiotti OE, Waples RS (2005) Assignment methods: matching biological questions with appropriate techniques. Trends Ecol Evol 20:136–142. doi:10.1016/j.tree.2004.12.004
Miller CR, Joyce P, Waits LP (2002) Assessing allelic dropout and genotype reliability using maximum likelihood. Genetics 160:357–366
Musiani M, Leonard JA, Cluff HD, Gates CC, Mariani S, Paquet PC et al (2007) Differentiation of tundra/taiga and boreal coniferous forest wolves: genetics, coat colour and association with migratory caribou. Mol Ecol 16:4149–4170. doi:10.1111/j.1365-294X.2007.03458.x
Neigel JE (1997) A comparison of alternative strategies for estimating gene flow from genetic markers. Annu Rev Ecol Syst 28:105–128. doi:10.1146/annurev.ecolsys.28.1.105
Ostrander EA, Sprague GF, Rine J (1993) Identification and characterization of dinucleotide repeat (ca)n markers for genetic mapping in dog. Genomics 16:207–213. doi:10.1006/geno.1993.1160
Packard J (2003) Wolf behavior: reproductive, social, and intelligent. In: Mech LD, Boitani L (eds) Wolves. Behavior, ecology, and conservation. University of Chicago Press, Chicago, pp 35–65
Paetkau D, Slade R, Burden M, Estoup A (2004) Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol 13:55–65. doi:10.1046/j.1365-294X.2004.02008.x
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855. doi:10.1046/j.1523-1739.1998.96489.x
Pilot M, Jedrzejewski W, Branicki W, Sidorovich VE, Jedrzejewska B, Stachura K et al (2007) Ecological factors influence population genetic structure of European grey wolves. Mol Ecol 15:4533–4553. doi:10.1111/j.1365-294X.2006.03110.x
Piry S, Alapetite A, Cornuet JM et al (2004) GeneClass2: a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539. doi:10.1093/jhered/esh074
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pulliainen E (1965) Studies on the wolves (Canis lupus L.) in Finland. Ann Zool Fenn 2:215–259
Pulliainen E (1980) The status, structure and behaviour of the wolf (Canis lupus L.) along the Fenno-Soviet border. Ann Zool Fenn 17:107–112
Räikkönen J, Bignert A, Mortensen P, Fernholm B (2006) Congenital defects in a highly inbred wild wolf population (Canis lupus). Mamm Biol 71:65–73. doi:10.1016/j.mambio.2005.12.002
Ramirez O, Altet L, Enseñat C, Vilà C, Sanchez A, Ruiz A (2006) Genetic assessment of the Iberian wolf Canis lupus signatus captive breeding program. Conserv Genet 7:861–878. doi:10.1007/s10592-006-9123-z
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9221. doi:10.1073/pnas.94.17.9197
Raymond M, Rousset F (1995a) An exact test for population differentiation. Evolution Int J Org Evolution 49:1280–1283. doi:10.2307/2410454
Raymond M, Rousset F (1995b) genepop (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Sacks BN, Mitchell BR, Williams CL, Ernest HB (2005) Coyote movements and social structure along a cryptic population genetic subdivision. Mol Ecol 14:1241–1249. doi:10.1111/j.1365-294X.2005.02473.x
Seddon JM (2005) Canid-specific primers for molecular sexing using tissue or non-invasive samples. Conserv Genet 6:147–149. doi:10.1007/s10592-004-7734-9
Seddon JM, Sundqvist A-K, Björnerfeldt S, Ellegren H (2006) Genetic identification of immigrants to the Scandinavian wolf population. Cons Gen 7:225–230. doi:10.1007/s10592-005-9001-0
Slatkin M (1985) Rare alleles as indicators of gene flow. Evolution Int J Org Evolution 39:53–65. doi:10.2307/2408516
Tallmon DA, Koyuk A, Luikart GA, Beaumont MA (2008) ONeSAMP: a program to estimate effective population size using approximate Bayesian computation. Mol Ecol Resour 8:299–301. doi:10.1111/j.1471-8286.2007.01997.x
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. doi:10.1111/j.1471-8286.2004.00684.x
Vilà C, Sundqvist A-K, Flagstad Ø, Seddon J, Björnerfeldt S, Kojola I et al (2003) Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc R Soc Lond B Biol Sci 270:91–97. doi:10.1098/rspb.2002.2184
Wabakken P, Sand H, Kojola I, Zimmermann B, Arnemo JM, Pedersen HC et al (2007) Multistage, long-range natal dispersal by a global positioning system-collared Scandinavian wolf. J Wildl Manage 71:1631–1634. doi:10.2193/2006-222
Waples RS (2006) A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7:167–184. doi:10.1007/s10592-005-9100-y
Waples RS, Do C (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756. doi:10.1111/j.1755-0998.2007.02061.x
Walsh PS, Metzger DA, Higuchi R (1991) Chelex® 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Weckworth BV, Talbot S, Sage GK, Person DK, Cooks J (2005) A signal for independent coastal and continental histories among North American wolves. Mol Ecol 14:917–930. doi:10.1111/j.1365-294X.2005.02461.x
Whitlock MC, McCauley DE (1999) Indirect measures of gene flow and migration: Fst ≠ 1/(4Nm + 1). Heredity 82:117–125. doi:10.1038/sj.hdy.6884960
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wolff JO (1997) Population regulation in mammals: an evolutionary perspective. J Anim Ecol 66:1–13. doi:10.2307/5959
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aspi, J., Roininen, E., Kiiskilä, J. et al. Genetic structure of the northwestern Russian wolf populations and gene flow between Russia and Finland. Conserv Genet 10, 815–826 (2009). https://doi.org/10.1007/s10592-008-9642-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-008-9642-x