Abstract
Data analysis workflows in High Energy Physics (HEP) read data written in the ROOT columnar format. Such data has traditionally been stored in files that are often read via the network from remote storage facilities, which represents a performance penalty especially for data processing workflows that are I/O bound. To address that issue, this paper presents a new caching mechanism, implemented in the I/O subsystem of ROOT, which is independent of the storage backend used to write the dataset. Notably, it can be used to leverage the speed of high-bandwidth, low-latency object stores. The performance of this caching approach is evaluated by running a real physics analysis on an Intel DAOS cluster, both on a single node and distributed on multiple nodes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The next years of the scientific programme at CERN will provide, among others, great challenges in storing and processing an ever larger amount of data coming from the Large Hadron Collider (LHC). The current roadmap highlights that by the next hardware update, named HL-LHC [5], both hardware and software improvements will need to happen to address future computing and storage needs [1, 21].
For a long time the particular use cases provided by High Energy Physics (HEP) data could only be approached by in-house solutions. This need has involved all aspects of the data lifecycle, from collection in the LHC, to storing the information of physics events, to running full scale analyses that very often comprehend both actual collider data and simulated events that are compared against each other. On the one hand, the high throughput with which data is generated during LHC runs (with peaks of a hundred PB for the last active period in 2018) has been addressed by multiple layers of skimming the raw data. The very first reduction is done at the edge of the collider components through hardware triggers that only save a fraction of the actual event information. This is then sent to storage in a so-called “raw” format, that is not directly representative of the actual physical interactions of the particles. All next steps involve further reduction of the data into different formats that differ in the on-disk representation of the particle information (from more complex to simpler). Finally, the skimmed datasets are used by researchers worldwide on a frequent basis (daily to weekly). The pipeline described, starting right after the first raw formats are modified to better represent physics events, is managed through a single software tool, ROOT [13]. This is a framework that has become the de facto standard in the field for storing, processing and visualising data in HEP. Many examples of its usage can be found within physics research groups for their analysis tasks [8, 49] as well as the basis for more complex software tools [2, 56].
When managing data, deciding its layout is crucial in understanding how the processing tools should work later. A physics dataset is comprised of events (which can be seen as rows of a table). Each event can have a complex structure, divided in multiple physical observables (which can be considered columns). The observables can hold simple objects (like integers or floating point numbers), but most often they are represented through nested structures (collections of collections of elements). Thus, the dataset schema has no direct SQL representation. While the number of rows is usually fixed (there are N events in a certain physical interaction), each column can store one value, an array of values or jagged arrays with no fixed relation between their sizes. ROOT defines a data layout (on disk and in memory) which is the format used when writing and reading all HEP datasets. It is binary, columnar and capable of storing any kind of user-defined object in a file. Since the software framework is mainly implemented in C++, a ROOT file can store arbitrary C++ objects, with an automatic compression mechanism that splits complex classes into simpler components. Also, using a columnar layout further reduces the amount of read transactions needed, since different columns can be read independently from each other. The underlying storage of such an efficient data format has traditionally been file-based only.
Although the information from the collider flows into multiple skimming steps as described above, the datasets used in HEP analyses can still represent a processing challenge. A full Run 2 dataset (with information taken between 2015 and 2018) can still hold multiple TB of information. ROOT has historically offered analysis interfaces that physicists worldwide have used for direct processing or to build more complex libraries. While providing an efficient way to process large datasets (also with sparse access thanks to their columnar layout), these interfaces were built with single-threaded sequential applications in mind. More recently, modern and more mature high-level interfaces have been introduced that rely upon the established structures while providing more performance through easy parallelisation on physicists’ machines. In particular, parallelisation is achieved through implicit multi-threading where each thread operates concurrently on a different portion of the input dataset. This exploits an important feature of HEP data: physics events are statistically independent. This means that processing the first hundred events or processing the last hundred events can happen completely in parallel. In this sense, HEP data analysis is an embarrassingly parallel problem.
In order to exploit this feature, parallel computing has always been extensively explored in this field, in particular in the form of grid computing. In this context the Worldwide Computing LHC Grid (WLCG) has served the computing needs of physicists all over the world for the last few decades [9]. The main technologies used to steer the distributed applications have been batch job queueing systems like HTCondor [47]. Due to the aforementioned file-based datasets and the fact that they are most often stored in remote facilities, distributed computing in HEP is inherently I/O bound. Batch jobs can often suffer from network instabilities and latency brought by geographical distance between computing nodes and storage facilities. To this end, different approaches at caching input data have been studied and employed in the past. Notably, the XRootD framework [20], which defines the standard protocol for remote data access used in HEP, also includes a file-based caching implementation. This is referred to as XCache by the physics community, and is used in various HEP computing facilities [22, 45].
All these efforts to bring efficiency into HEP data processing workflows notwithstanding, future challenges can and should also be addressed by looking at possible alternative approaches. For example, a more recent approach to data storage has been presented by object store technologies. These technologies are widely used in cloud-based applications or in HPC facilities, which are built to support highly scalable workflows [17, 31, 32, 46]. Literature shows that object stores are able to overcome some limitations of traditional POSIX file-based systems and provide efficient data access in distributed environments. An example of this can be found in a work by Liu et al. [28], where a benchmark of parallel I/O comparing different object stores with the Lustre filesystem [12] demonstrated that the latter suffers from POSIX constraints and filesystem locking overhead when scaling to more processes on one node.
This work describes a novel approach at improving performance of real physics analyses by reading the input data from an object store, rather than from files. Since HEP analyses are usually I/O bound, the goal is to study how a high-bandwidth low-latency object store can help speed up such analyses. For this purpose, the ROOT I/O system is extended with an automatic caching mechanism so that, during the execution of an analysis, the input data that is read from remote files is cached in an object store, with the goal of reusing that cache in subsequent (faster) executions on the same dataset. An example of a HEP analysis application published by the LHCb experiment [50] is used as benchmark, both in a single-node and multi node scenario.
The rest of the paper is structured as follows. Section 2 highlights the current status of this type of research in the HEP field, as well as mentioning similar approaches in industry. Section 3 discusses the software tools employed for the purposes of this study. Section 4 gives more details about the proposed solution to tackle the challenges described above. Section 5 highlights the tests created and run in order to test the validity of the proposal; the test results are presented and discussed. Section 6 summarises the achievements of the paper.
2 Related work
Object storage technology is typically used in HPC and distributed computing scenarios. Over the years, various implementations of this system have spawned within industry. Notable examples are provided by vendors such as Amazon with the S3 service [3], Microsoft with the Azure Blob Storage [14] and Intel with the Distributed Asynchronous Object Store (DAOS) [27]. All of them usually suppose a system of nodes which, while being distributed, is highly coherent and localised. For example, computing and storage nodes usually belong to the same network which is also often cut off from the internet. This approach may lack the flexibility required in other distributed computing fields such as the Internet of Things (IoT). One of the biggest issues with data caching in this context are content redundancy and cache overflow, which also apply to the HEP context. For example, different physics research groups may be interested in the same datasets and a reasonable caching strategy must make sure that the large physics datasets are not copied over multiple nodes if they are already available. These topics are not yet explored in the HEP field, but literature already shows that a good bandwidth utilization can be achieved given a large enough cache size on network nodes [19], which is the usual situation in HEP storage facilities.
There is literature comparing different technologies according to established benchmark suites [25, 28]. In some cases, current knowledge allows to extract the best performance of a given object storage tool, through fine-tuning of user space parameters [42]. This work does not attempt to modify or tune the storage backend; rather, it focuses on addressing analysis needs from the perspective of the data format and the layer that implements I/O of the data format to various backends. There are other examples of I/O libraries that have attempted an integration of their data format with fast object stores, such as the HDF5 connector for Intel DAOS [44].
Regarding the execution of distributed workflows that exploit object stores, some efforts can be found for industry products [40, 41, 51]. In the cited approaches, the object store is used as as scalable storage layer to host big datasets and the computing nodes read data directly from the object store. Furthermore, it is shown that once the object store semantics are leveraged properly, read-intensive analysis workflows can get 3-6 times faster.
Regarding caching large input datasets, very rarely do other investigations highlight the possible benefits that it could have in distributed computing scenarios. In a work that compared different object store engines in geospatial data analysis workflows [43], a part of the benchmark presented the improvements in performance of the different engines with caching. But only the filesystem cache was used in that case, hence data and queries could be kept in the memory of the nodes and no separate caching mechanism was implemented.
In the HEP context, object stores still do not see widespread usage, even in large scale collaborations. Research studies from the early years of the LHC have tried integrating object stores in the grid through the Storage Resource Manager (SRM) interface [7]. This interface allowed accessing and managing storage resources on the grid. In a first effort, a plugin was developed to connect Amazon S3 resources to the grid storage layer [4]. Later on, a tier 2 grid facility of the ATLAS LHC experiment [48] was extended to use the Lustre filesystem, with benchmarks showing 8 GB/s of peak read speed [55]; this work is able to reach a much higher throughput, as will be shown in Sect. 5. More recently, the focus has migrated towards the evaluation of such storage solutions on concrete examples of HEP software like ROOT [6], where a physics dataset was used to evaluate data access patterns over an S3 API, leading to aggregated throughput of a few Gigabytes per second. An interesting example of investigation into data analysis needs can be found in a work by Charbonneau et al. [16], where 8 TB of physics events are stored in a Lustre cluster; this work reveals that although resource scaling helps achieve higher throughput, remote data access while processing can become a burden for analysts. It is clear from this few examples that the potential of object stores, especially newer approaches that rely on low-latency high-bandwidth systems like DAOS, has not been extensively explored in this field. In fact, even very recent mentions of such systems are still a topic of discussion in internal workshops at CERN [15, 29].
The focus of this work is how a caching layer implemented on top of a fast object storage connector could bring HEP data analysis use cases a tangible speedup when reading ROOT data. Per the review of the field literature available, no such evaluation has been previously addressed. The main missing point of other approaches is the focus on the specific data format, and how it can be ingested in an object store for efficient querying of any needed subset of dataset properties. A previous article compared the behaviour of already existing caching mechanisms in HEP software with respect to two configurations: caching in the computing nodes or in a dedicated cache server [34]. That publication showed limitations of existing software when interfacing to the traditional file-based ROOT I/O. A more recent effort developed an integration between the DAOS C API and the next generation ROOT storage layer, demonstrating its flexibility in terms of different storage backends it can support [30]. This work relies on the findings of these two investigations and includes the following novel contributions:
-
A caching mechanism is implemented in C++ and plugged in the ROOT I/O layer. It is independent of the storage backend, so that a ROOT dataset can be opened from a remote file-based server and the cache can be stored on a fast object store.
-
The performance of a physics analysis that reads a dataset already cached in an object store is measured not only in single-node, but also in multi-node runs.
-
To support the latter use case, the RDataFrame analysis library of ROOT was extended in this work to be able to read data from a DAOS object store. The previous implementation only supported reading the traditional file-based data format.
3 Background
A few key software tools have been employed for the purposes of this investigation. Since the caching mechanism is developed directly at the ROOT I/O level, it relies on low-level primitives to open, read and write into a ROOT file. Furthermore, DAOS is the storage backend used in the benchmarks of Sect. 5. Finally, the main objective is to give analysis tools a boost in read performance of input data, so this work relies on the high-level modern interface for data analysis offered in ROOT. This section describes in more detail these three components, highlighting their specific relevance to this study.
3.1 Intel DAOS
The object store chosen for the purposes of this work is Intel DAOS [27], a fault-tolerant distributed object store targeting high bandwidth, low latency, and high I/O operations per second (IOPS). DAOS addresses traditional POSIX I/O limitations on two fronts in order to optimise data access. On the one hand, it bypasses kernel I/O scheduling strategies, e.g. coalescing and buffering, that are mostly relevant for high-latency few-IOPS spinning disks. On the other hand, it avoids using the virtual filesystem layer, since the strong consistency model enforced by POSIX is known to be a limiting factor in the scalability of parallel filesystems.
On a DAOS system, there are two different categories of nodes: servers and clients. All data in DAOS is stored on the server nodes. There can be many servers running a Linux daemon that exports local NVMe/SCM storage. This daemon listens on a management interface and several fabric endpoints for bulk data transfers. RDMA is used where available, e.g. over InfiniBand [35] or OmniPath [10] fabrics, to copy data from servers to clients. The client nodes are the ones responsible for running computations defined by the users in their applications. A DAOS client node does not store any data on itself, rather it requests the dataset from the servers when it’s needed.
The storage is partitioned into pools and containers that can be referenced by means of a Universally Unique IDentifier (UUID) [23]. Objects can be partially read or written into a container. Each of these objects is a key-value store that is accessed using a 128-bit Object IDentifier (OID). Object data may additionally have redundancy or replication.
Data layout of RNTuple [11]
3.2 ROOT I/O
It has been mentioned that ROOT is the standard software used by physicists around the world for all their needs revolving around storing, processing, visualising data. In fact, one of the key components in ROOT is the I/O subsystem, which is were the data format described in Sect. 1 is defined. Traditionally, the I/O layer in ROOT was implemented in the TTree class [39], and more recently in RNTuple [38].
A TTree is a generic container of data, capable of holding any type of C++ object. This goes from fundamental types to arbitrarily nested collections of user-defined classes. TTree organises data into columns, called branches. A branch can contain complete objects of a given class or be split up into sub-branches containing individual members of the original object. Each branch stores its data in one or more associated buffers on disk. Different branches can be read independently, making TTree a truly columnar data format implementation. The implementation of TTree assumes that the underlying storage backend is file-based and it does not support I/O to object stores in any way.
The ROOT I/O subsystem is able to read and write datasets both to a local disk on the computer and to remote machines with protocols such as HTTP or XRootD [20]. The latter is the most common protocol for remote data access used in the HEP field. This makes ROOT datasets easily transferable from one physicist’s machine to another’s for easy sharing among colleagues or from large storage facilities to the various computing nodes that may be used to run production analyses.
TTree has been successfully used in the past to efficiently store more than one exabyte of HEP data and has become the de-facto standard format in the area. Its on-disk columnar layout allows for efficient reading of a set of selected columns, a common case in HEP analyses. However, future experiments at the HL-LHC are expected to generate one order of magnitude larger datasets which makes researching the benefits of using high-bandwidth low-latency distributed object stores especially relevant. As previously mentioned, TTree only supports I/O transactions to file-based systems so it does not provide the flexibility needed for future challenges.
RNTuple [11] is the new, experimental ROOT columnar I/O subsystem that addresses TTree’s shortcomings and delivers a high read throughput on modern hardware. In RNTuple data is stored column-wise on disk, similarly to TTree and Apache Parquet [54]. An overview of the data layout design is depicted in Fig. 1.
Specifically, data is organized into pages and clusters: pages contain values for a given column, whereas clusters contain all the pages for a range of rows. The RNTuple meta-data are stored in a header and a footer directly within the RNTuple object. The header contains the schema of the RNTuple; the footer contains the locations of the pages. The pages, header and footer do not necessarily need to be written consecutively in a single file. As long as the target container of the RNTuple specifies the location of header and footer, data can be stored in separate containers (e.g. different files or different objects in an object store).
The RNTuple class design comprises four decoupled layers. The event iteration layer provides the user-facing interfaces to read and write events and can be used from higher-level components in ROOT such as RDataFrame, which is described in Sect. 3.3. The logical layer defines the mappings to split arbitrarily complex C++ objects into different columns of fundamental types. The primitives layer manages deserialised pages in memory and the representation of fundamental types on disk.
RNTuple’s layered design decouples data representation from raw storage of pages and clusters, therefore making it possible to implement backends for different storage systems, such as POSIX files or object stores. Recently, a DAOS backend for RNTuple was developed and demonstrated promising performance results [30]. At the time of writing, this backend uses a unique DAOS OID to store data for each page.
3.3 ROOT RDataFrame
RDataFrame is the high-level interface to data analysis offered by ROOT [36]. It features a programming model where the user calls lazy operations on the dataset through the API and the tool effectively builds a computation graph that is only triggered when the results are actually requested in the application. This interface supports processing of traditional TTree datasets but also other data formats, among which RNTuple.
Parallelisation is a key ingredient in an RDataFrame workflow. The native C++ implementation allows to use all the cores in a single machine through implicit multithreading. Furthermore, RDataFrame computations can be distributed to multiple nodes through its Python bindings [33]. The design of the distributed RDataFrame extension accommodates multiple backends (schedulers); the application code does not vary when moving from one backend to another.
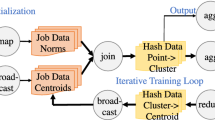
Overview of the proposed system. The upper box includes the main ROOT components involved in an analysis. On the left of the dashed line (Analysis layer) is the user-facing API and the processing engine offered by RDataFrame. On the right is the I/O layer that brings compressed physics data from disk to uncompressed information in memory that is sent to RDataFrame for processing. Contrary to the traditional ROOT I/O layer implemented with TTree, this work focuses on the next-generation system implemented with RNTuple. The two orange boxes represent the parts introduced in this work: the introduction of RNTuple as a supported input data format for the distributed RDataFrame layer in the Analysis layer and a caching mechanism for RNTuple in the I/O layer
4 Design of the caching system
The context exposed in Sect. 1 highlights an important aspect of HEP data analysis that should be addressed, namely data access at runtime. HEP analysis is often I/O bound, either because the datasets reside in remote locations or because there is little computation involved. In such scenarios, it is greatly beneficial to enable caching of the input datasets, storing them as close as possible to the computing nodes. This calls for a new solution that exploits the future storage layer provided by RNTuple.
Thus, this work focuses on evaluating the benefits of using a novel storage architecture, such as the one offered by DAOS, when reading input data of a HEP analysis. For that purpose, a caching machinery has been developed for data reading via RNTuple, so that any application that reads RNTuple data could benefit from it. This includes notably RDataFrame applications that read data from RNTuple. The caching mechanism is independent of the storage backend, a crucial feature to maintain transparency for the user and contribute towards a sustainable development in a future where RNTuple will be able to read and write to even more storage systems than today.
Figure 2 gives a high-level view of the interaction between ROOT and DAOS after the proposed developments. A physics analysis with ROOT makes use of two main components: an analysis layer and an I/O layer. The main analysis interface in ROOT is RDataFrame, which offers a declarative user-facing API and a lazy execution engine as described in Sect. 3.3. The user provides a dataset specification to RDataFrame, for example a list of files to process. In turn, RDataFrame will transparently invoke the low-level I/O layer which is in charge of opening the files from disk, uncompressing their data and sending them back to the processing layer. The image shows in particular the I/O layers defined within RNTuple as described in Sect. 3.2. All the blue boxes in the figure represent already established ROOT components, while the orange boxes demonstrate the parts that were specifically modified or developed in this work. In particular, the distributed RDataFrame layer was not able to process data coming from the RNTuple I/O. After this work, the algorithm that creates a distributed RDataFrame task on the client node also checks the origin of the dataset. This allows creating the correct RNTuple object when the task arrives on the computing node (bottom left part of the image). When a distributed task starts executing, it will create an RNTuple instance to read data from the selected storage. In case the RNtuple cache is activated, this can transparently start writing data from the original storage system to the target one. For the purposes of this work, the target storage system for the RNTuple cache is DAOS. If the DAOS server already contains the desired dataset, the developed cache will serve it directly to the rest of the RNTuple I/O pipeline which will in turn direct it towards the RDataFrame that requested it inside the distributed task.
Schema of the newly developed caching system in the RNTuple I/O pipeline. Blue horizontal arrows represent the current two steps of the pipeline: reading compressed pages and decompressing them. a represents the case where the application is reading from some file-based source and a new RNTuple object is created to write data to a cache. In (b), data is read from the cache during the analysis (Color figure online)
4.1 Integration within RNTuple
RNTuple I/O operations are scheduled in a pipeline. The current implementation of the pipeline is in two steps: first, data is read from storage into compressed pages in memory, then bunches of pages are decompressed together and sent to the rest of the application for processing. The caching mechanism takes place in between the two steps of the pipeline. This can be described as follows:
-
1.
When programming an application, the user can enable the cache by simply providing a storage path (to a local directory or to an object store address for example) as an extra option when opening an RNTuple.
-
2.
In order to write the cache, a new RNTuple is created with the same metadata as the original RNTuple, this time pointing to the storage path provided by the user in step 1. Figure 3a shows the input dataset to the left, in red. The metadata, i.e. the list of three column names, are mirrored in the RNTuple cache that is shown on the right side of the image.
-
3.
At a later stage, when the first step of the RNTuple pipeline is over, the compressed pages read into memory are grouped together in a cluster object. The cluster object contains a list of column names that the cluster is spanning. From any column name, a group of (compressed) pages belonging to that column can be retrieved. Consequently, the caching algorithm proceeds by traversing all column names and for each column it writes the corresponding pages into the newly created RNTuple object, thus populating the cache location (see Fig. 3a). It is important to note here that the RNTuple system is implemented such that the column metadata is stored separately from the actual compressed pages (or groups thereof), so that information needed in the I/O pipeline is always available.
-
4.
When the reading part is over, the RNTuple cache object finalises the writing operations and closes the open handle to the storage path (e.g. writes metadata about the number of pages and cluster layout to an attribute key in the DAOS case).
-
5.
Any subsequent access to the same dataset by the user, will fetch the cached RNTuple rather than the original one. The caching mechanism is completely bypassed in order to avoid extra operations and the user is transparently presented with an RNTuple that resembles exactly their input dataset, but is read from a fast storage system like DAOS (Fig. 3b).
4.2 Optimisations for HEP use cases
Another important notion to discuss revolves around how the cache will interact with the ROOT I/O layer. As discussed in Sects. 1 and 2, the data lifecycle in HEP is such that big collaborations at the LHC gather data from collisions in the accelerator when it is functioning, writing it into storage facilities. Once written, this information is set in stone. On top of that, many simulations can be done over the years if new or more precise models arise to be checked against the real data. Running an experiment simulation campaign represents possibly the only other situation where storage facilities are hit with a large amount of write operations. In any case, HEP data is characterised by a “write-once, read-many” condition. Consequently, any caching system aimed at analysts’ needs in this field will optimize read operations as much as possible. Writing is also important to smoothen the user experience when the cache is still cold, but providing a faster reading performance directly translates into an increased productivity in physics analysis research. The machinery developed in this work tries to address both objectives: when writing, the cache does not need to wait for the decompression step of the RNTuple pipeline; when reading, it directly forwards all requests to low-level efficient RNTuple interfaces.
4.3 Interaction of the caching system with DAOS
The I/O workflow from the point of view of the caching node (which in this work corresponds to a DAOS server) looks like this: the user starts an analysis, requesting to process some dataset; the dataset is opened and both sent from disk to memory for processing and at the same time written as-is into the target caching node; after this, any other time an analysis is run and requests the same dataset, it is automatically read from the cache. Overall, the number of read operations is much higher than the number of write operations in this context.
The DAOS specification also establishes the use of caches to boost data access for its users. This is implemented at various levels, for example it is always enabled by default in case the dfuse layer is used [18]. At the hardware level, it exploits burst buffers on the server nodes [27]. All of these characteristics are completely transparent and orthogonal to the caching system for RNTuple developed in this work. This is, from the point of view of DAOS, just like any other user application that reads or writes data stored in the DAOS servers. Thus, any improvement to the DAOS library or any site-specific tuning enabled on the server nodes will automatically be leveraged by the RNTuple cache.
HEP data analysis is most often I/O bound, due to having to read very large datasets usually stored remotely. A certain physics analysis may require to read only a subset of the available columns in the dataset. This is made possible thanks to the columnar layout of the ROOT data format. For the purposes of this study, Sect. 5 shows an example of real HEP analysis where only some columns are read, thus also demonstrating the possibility of caching an RNTuple resulting from accessing a fraction of the input dataset.
This paper focuses only on the point of view of a single user. In a multi-user scenario, this caching system embedded in RNTuple should be synchronised with a storage-facility-wide service. Taking for example two different users that want to access the same dataset, whoever does access it first will cache it in the object store thanks to the system developed in this work. But in order for the other user application to know about the presence of the dataset in the cache, some dataset register should be queried and report whether the same data is already present. This kind of challenge will be topic of further studies.
5 Experiments
This section will present various test configurations that were employed to evaluate the capabilities of the proposed caching mechanism. At first, the cache is exercised on a small dataset, without running a physics analysis but just comparing the reading speed of the RNTuple cache on DAOS with the reading speed from a local SSD. Afterwards, a real HEP analysis is performed with the RDataFrame tool, either on one node or distributed to multiple nodes. For this second type of test, two different clusters have been used. The first cluster features a DAOS system where the RNTuple cache can store data on the DAOS servers and send it to the RDataFrame engine for processing. The second cluster has a Lustre shared filesystem and in this case the same distributed RDataFrame analysis processes data with the traditional ROOT I/O system using TTree.
5.1 Testbed specification
5.1.1 DAOS cluster
In the DAOS cluster there are seven client nodes and two servers. According to the DAOS specification, the dataset that is processed in the experiments described in the next sections is always stored on the server nodes. Specifically, the server nodes are the caching nodes. Client nodes on the other hand only read the data from the server nodes and run the computations defined in the physics analysis. Each client node features the following hardware specifications:
-
Motherboard: Newisys DoubleDiamond TCA-00638.
-
CPU: 2x Intel Xeon E5-2640v3, for a total of 2 NUMA sockets, 8 physical cores per socket, 2 threads per core.
-
RAM: eight Micron 36ASF2G72PZ-2G1A2 DIMMs, 16 GiB each, for 128 GB of total memory.
-
Inifiniband interfaces: one Mellanox MCX354A-FCBT two port NIC, only one port was cabled, 56 Gb/s FDR speed; one HPE P23842-001 two-port NIC, only one port was cabled, 100 Gb/s EDR speed. Each interface is connected separately to one NUMA socket.
Each server node features the following hardware specifications:
-
Motherboard: Supermicro X11DPU-Z+.
-
CPU: 2x Intel Xeon Gold 6240, for a total of 2 NUMA sockets, 18 physical cores per socket, 2 threads per core.
-
RAM: twelve Hynix HMA82GR7CJR8N-WM volatile DIMMs, 16 GB each, 6 per socket. 192 GB total memory.
-
DAOS storage: twelve Intel HMA82GR7CJR8N-WM persistent memory DIMMs, 128 GB each, 6 per socket. Also, eight Samsung MZWLJ3T8HBLS-00007 3.84 TB NVMe SSD, four per socket.
-
Infiniband interfaces: two Mellanox MCX654105A-HCAT one-port NICs 200 Gb/s (HDR). Each interface is connected to one NUMA socket. The node has PCIe Gen3 buses, so the actual bandwidth is 100 Gb/s per NUMA socket.
In practice, the maximum bandwidth that can be obtained when reading data on one of the client nodes is given by the sum of the two nominal bandwidths of its Infiniband interfaces. That is, a client node can read up to 156 Gb/s, or 19.5 GB/s.
The maximum bandwidth overall of the whole DAOS cluster is given by the sum of the nominal bandwidths of the server nodes. Thus the maximum reading throughput for the whole cluster is 400 Gb/s or 50 GB/s.
The DAOS version installed on this cluster is 1.2.
5.1.2 Lustre cluster
The second cluster used for this work is a large computing cluster with hundreds of nodes and a shared Lustre filesystem. Access to the cluster was granted via a user account registered with the Slurm resource manager [24] of the cluster. Cluster resources were shared among many other users. Also in this case there are server nodes where data is stored (on Lustre) and computing nodes that read the data from the server nodes and run the computations.
Each client node features the following specifications:
-
Motherboard: Supermicro H11DST-B.
-
CPU: 2x AMD EPYC 7551, for a total of 2 NUMA sockets, 32 physical cores per socket, 2 threads per core.
-
Inifiniband interface: one Mellanox ConnectX-4 VPI adapter card, FDR IB 40GbE, 56 Gb/s.
The network topology is built like a fat-tree, with a 2 to 1 blocking factor on the computing nodes. More information about this cluster can be found in its user manual [53].
5.2 Methodology
The following groups of tests have been set up for the caching system developed in this work:
-
1.
A single-threaded C++ application that uses the RNTuple interface to read sequentially all the entries of a dataset stored in a ROOT file. No other computation is done in the application. The dataset size is 1.57 GB. The purpose of this test is comparing the runtimes of three different configurations: (i) reading the dataset stored in a file on the local SSD of a node; (ii) reading the dataset stored in a file on the local SSD of a node and at the same time caching data to DAOS; (iii) reading the dataset stored in the DAOS cache.
-
2.
An open data analysis of the LHCb experiment at CERN [26], named from here on “LHCb benchmark”. This analysis is run on both clusters described in Sect. 5.1. It uses the distributed RDataFrame tool to steer computations from one to multiple nodes. In the DAOS cluster, it reads data from the DAOS servers through the RNTuple cache. In the Lustre cluster, I/O is done with the traditional TTree implementation. The application processes the dataset used in the cited publication, replicated eight-hundred-fold to get a 1259 GB sample. In the tests with TTree and Lustre, the dataset is replicated simply by providing a list of paths to multiple copies of the original dataset. In the tests with RNTuple and DAOS, the replicated dataset is obtained by running a C++ program that reads all the entries in the original file (stored on the same SSD of the node in the previous group of tests) and copies them to one or more separate RNTuple objects stored in DAOS, until the desired size is reached. The number of objects is equal to the number of distributed RDataFrame tasks, so that each task processes exactly one RNTuple object. As previously discussed, particle physics events are statistically independent, so this approach is valid for benchmarking purposes.
The benchmarks in Sect. 5.3 request a variable amount of nodes and cores per node on the cluster through the distributed RDataFrame tool. In all tests, 2 GB of RAM are requested per core. For the DAOS tests, data is always cached on the DAOS server nodes, never on the computing nodes. The tests on the Lustre cluster present a similar situation.
The tests done on the cluster with the Lustre filesystem are run by submitting jobs to the Slurm resource manager. In each job, the desired number of nodes and cores for that test is requested. Furthermore, each job requests exclusive access to all the computing nodes involved in the test, to avoid unpredictable loads on the machines due to shared usage with other users of the cluster.
The test suite is available in a public code repository [52].
5.3 Results
5.3.1 Caching RNTuple to DAOS
The first group of tests described in Sect. 5.2 is executed on a single node. The dataset is initially stored on SSD in order to gather more consistent measures and avoid possible network instabilities. Nonetheless, the same tests could be repeated with the dataset stored in a remote file, since RNTuple data can be also read through HTTP.
Table 1 shows average runtime of the application with three different configurations. On the one hand, caching to DAOS while reading from SSD brings roughly 50% overhead with respect to only reading from SSD. On the other hand, reading from the DAOS cache is more than 6 times faster than reading from SSD.
Processing throughput (i.e. reading the dataset and running analysis computations on it) of a distributed RDataFrame analysis on a single node of the DAOS cluster. a Real throughput values compared with a linear throughput increase obtained by multiplying the throughput on one core by the number of cores on the x axis. b Speedup obtained by scaling the analysis to multiple cores on the node
Processing throughput (i.e. reading the dataset and running analysis computations on it) of a distributed RDataFrame analysis on multiple nodes of the DAOS cluster (using 16 cores per node). a Real throughput values compared with a linear throughput increase obtained by multiplying the throughput on one node by the number of nodes on the x axis. b Speedup obtained by scaling the analysis to multiple nodes of the cluster
5.3.2 Distributed RDataFrame analysis benchmarks reading data from DAOS
A second series of tests evaluate the performance of running an RDataFrame analysis on top of RNtuple data cached in DAOS. The LHCb benchmark described in Sect. 5.2 is executed in a Python application with the distributed RDataFrame tool. This allows to parallelise the analysis both on all the cores of a single machine and on multiple nodes, all with the same application. Furthermore, while the user code is written in Python, this is just used as an interface language and each task is actually running C++ computations through RDataFrame. Within the test, the dataset is split into multiple RNTuple objects stored in DAOS. Then, one task is defined to run the analysis on a single RNTuple in its own Python process. In general, for any given number of cores used in the following tests, there are as many Python processes and as many RNTuple objects stored in DAOS.
At runtime, the application is monitored with a timer that is used to compute the processing throughput (that includes time spent reading and time spent in the computations). 72% of the total dataset is read and processed, roughly 904 GB. The processing throughput is then computed dividing the processed dataset size by the execution time. Figures 4 and 5 both show the absolute value of the processing throughput (with increasing amount of cores either with a single node or multiple nodes) and the value relative to one core for the single-node case or one node for the multi-node case.
The following results are representative of tests where the application processes are pinned to run on either NUMA domain of the node. The backend of distributed RDataFrame is set up such that there are two executor services running on the node, one that will accept and process tasks running on the first NUMA domain, the other running its tasks on the second NUMA domain.
Figure 4 shows the throughput obtained by running the application on one node, with an increasing amount of cores up to 16 (8 physical cores per NUMA domain). In particular, Fig. 4a shows the absolute processing throughput on a single node with increasing amount of cores. Here it can be seen that this tool is able to reach a peak processing throughput of more than 8 GB/s on one computing node. Figure 4b instead reports relative speedup on one node, which in this case is almost perfectly linear. In both images, up until 8 cores the test is using one of the NUMA domains on the node. When more than 8 cores are used, 8 of them are pinned to run on the first NUMA domain of the node, while the remaining are pinned on the second NUMA domain, to factor out NUMA effects.
The same analysis is then scaled to multiple nodes. In this scenario, presented in Fig. 5a, the peak processing throughput achieved is 37 GB/s, while the speedup plot in Fig. 5b shows a plateau when more than five nodes are being used.
Processing throughput (i.e. reading the dataset and running analysis computations on it) of a distributed RDataFrame analysis on multiple nodes of the cluster with the Lustre filesystem (using 16 cores per node). a Real throughput values compared with a linear throughput increase obtained by multiplying the throughput on one node by the number of nodes on the x axis. b Speedup obtained by scaling the analysis to multiple nodes of the cluster
5.3.3 Distributed RDataFrame analysis benchmarks reading data from Lustre
The same physics analysis is then run on the cluster that uses the Lustre shared filesystem for data access. In this case, the traditional ROOT I/O system using TTree is put to the test with a file-based storage system. The LHCb benchmark described in Sect. 5.2 is executed in a Python application with the distributed RDataFrame tool. The same dataset with the same size described in Sect. 5.2 is processed, with the only difference being that it is stored in the TTree data format instead of RNTuple.
One other difference in this case is that the TTree I/O, being more mature than RNTuple, already implements a way to read only a group of rows from a certain dataset when requested. Thus, the distributed RDataFrame tool is already capable of automatically splitting the user-provided dataset specification (i.e. the list of files to be processed) into multiple tasks, each containing a range of entries to process. When a task reaches a computing node, it will automatically create a local RDataFrame and open a TTree-based dataset reading only the entries supplied in the task metadata.
Figure 6 shows the throughput obtained by running the application on an increasing number of nodes of the cluster, in order to recreate as closely as possible the same configuration used in the tests described in Sect. 5.3.2. In particular, from one to seven computing nodes are requested to the Slurm resource manager, with exclusive access in order to avoid unpredictable CPU load from other users of the cluster. On each node, the benchmark requests exactly 16 physical cores. Each core will be assigned with one task, that will read a portion of the dataset as described above from the Lustre filesystem.
In Fig. 6a, the processing throughput obtained on an average of 10 benchmark runs per node count is compared with a linear throughput increase obtained by multiplying the value at the 1-node mark by the number of nodes on the x axis. The maximum throughput achieved is 13.3 GB/s. Figure 6b reports the speedup of running the analysis on multiple nodes relative to one node, comparing it with a linear speedup. The figure shows a perfect alignment between the two lines until the 4-node count, with a slight decrease in real speedup when using more nodes.
5.4 Discussion
Adding a new caching mechanism to a complex library such as ROOT requires careful design, both for usability and performance purposes. The proposed design is completely transparent to the user, who still only has to program their analysis through the RDataFrame high-level API. Drawing inspiration from the flexibility offered by the RNTuple layers described in Sect. 3, the caching system is injected in the I/O pipeline and can run completely in parallel with respect to the other operations issued to the underlying storage. The developed cache is backend-independent, thus enabling reading and writing RNTuple objects from/to any of the supported storage backends. This approach is the most sustainable in a field such as HEP, where large datasets can be stored in many different facilities around the world, each one with their own storage architecture.
In this work, the cache was exercised at different levels. At first, a physics dataset was either stored on an SSD or cached to DAOS with the developed tool. The results in Table 1 are promising for the caching mechanism. When caching data that is being read from the local SSD of a node, it is expected to have some overhead. But the speed gained when reading from the DAOS cache more than compensates this overhead. It is also worth highlighting that the main use case for such a tool is when the dataset is still in a remote location. In fact, the same dataset used in these tests and the following ones was originally stored at CERN and downloaded locally on the cluster. The time to download the dataset was not included in Table 1, but it is safe to state that long-distance network I/O does not achieve the same reading speeds as a local SSD.
The following results presented in Sect. 5.3.2 demonstrate the capabilities of the distributed RDataFrame tool used in conjunction with the new storage layer offered by RNTuple and its DAOS backend. In this case the dataset was replicated to reach a size of more than 1 TB, in order to give enough workload to the computing nodes. This was chosen in accordance with the usual dataset size for an LHC Run 2 analysis, which is in the order of one to a few tens of Terabytes.
On the DAOS cluster, the analysis reached a peak processing speed value of 8 GB/s and 37 GB/s respectively with one node and seven nodes. Comparing these numbers with the maximum bandwidths described in Sect. 5.1.1 reveals that the peak processing throughput on one node is equal to 40% of the maximum reading throughput, while the peak processing throughput on seven nodes is equal to 74% of the maximum reading throughput of the whole cluster. It must be noted that the processing throughput numbers include all the steps of the analysis: opening the RNTuple objects, reading the data, bringing it to the RDataFrame processing layer and running the computations defined in the analysis application. Thus, this first result is promising considering that the HEP analysis are I/O bound. The scaling shown is close to ideal on one node with processes pinned to either NUMA domain, less than ideal when using multiple nodes. In light of these results, it appears that the current implementation of the RNTuple DAOS backend needs to be further improved in order to saturate the full bandwidth of the cluster.
The results obtained on the DAOS cluster can be compared with the benchmarks shown in Sect. 5.3.3, which involve running the same analysis on another cluster that uses the Lustre filesystem. The comparison, while not done on exactly the same hardware because the DAOS cluster does not provide a Lustre filesystem, is still representative of the advantages offered to ROOT by a low-latency high-bandwidth object store like DAOS. The Lustre cluster also uses Infiniband interfaces like the DAOS cluster. All the same, the results presented in Fig. 6 show an overall worse performance of the analysis. It should be considered that the slope of the speedup is better in this case, almost aligned to a linear speedup all the way up to seven nodes. This is due to TTree being a much more mature I/O system than RNTuple, especially considering the RNTuple interaction with DAOS. In fact, file-based I/O in ROOT dates back to its very beginning in 1995. In spite of this difference in maturity between the two I/O systems, RNTuple with DAOS is able to achieve an almost three times higher processing throughput than TTree with Lustre at the moment. This demonstrates the potential gain of exploiting a high-throughput system such as DAOS as a data source for HEP analysis in ROOT with RNtuple, compared with a traditional file-based approach with TTree, even when the latter is supported by a first-class parallel filesystem that is currently used by most of the top supercomputers in the world [37].
6 Conclusions
This paper has demonstrated how object stores can be used to speed up real HEP analysis use cases, achieving an unprecedented processing throughput in single-node and multi-node parallel analysis execution.
A first native caching mechanism for the future I/O system in ROOT has been developed. The machinery is independent of the storage backend used, so that it is possible to run an application that transparently reads a remote dataset from a POSIX filesystem and writes it to an object store. This opens the door to previously unavailable fast storage systems to cache HEP data in an analysis environment. Furthermore, the generality of this machinery paired with the efficient design of RNTuple could potentially cater to data storage needs of other fields and use cases.
The caching system was put to the test in a fast object store such as Intel DAOS. When running a simple application that reads a remote file and caches it to DAOS, the overhead present while caching the dataset the first time it is being read is highly compensated by the much faster read speed obtained with DAOS. When pairing RNTuple with the RDataFrame analysis interface, very high read and processing throughput values were achieved on one node. Distributed RDataFrame allowed making better use of the available cluster resources, reaching a peak processing throughput of 37 GB/s on seven nodes (16 cores per node). A comparison was performed by testing the selected physics analysis with distributed RDataFrame processing a TTree dataset stored on a Lustre filesystem, using the traditional ROOT I/O system. This resulted in a 2.8 times lower processing throughput, peaking at 13.3 GB/s with the same number of nodes and cores used in the DAOS benchmarks.
Data availability
The original physics analysis used in the benchmarks shown in Sect. 5 is available at http://opendata.cern.ch/record/4902 and its related dataset at http://opendata.cern.ch/record/4900. All the code to augment the original dataset, process the benchmarks and visualize the results is available in the public repository: https://github.com/vepadulano/rdf-rntuple-daos-tests. The implementation of the caching system demonstrated in this work is available at https://github.com/vepadulano/root/tree/rntuple-cache-release-v2.
References
Aleksa, M., Blomer, J., Cure, B., et al.: Strategic R &D Programme on Technologies for Future Experiments. Tech. rep, CERN, Geneva (2018)
Altenmüller, K., Cebrián, S., Dafni, T., et al.: REST-for-Physics, a ROOT-based framework for event oriented data analysis and combined Monte Carlo response. Comput. Phys. Commun. 273(108), 281 (2022). https://doi.org/10.1016/j.cpc.2021.108281
Amazon Amazon Simple Storage Service Documentation. https://docs.aws.amazon.com/s3/. Accessed 1 Feb 2022 (2021)
Andreozzi, S., Magnoni, L., Zappi, R.: Towards the integration of StoRM on Amazon Simple Storage Service (S3). J. Phys. 119(6), 062011 (2008). https://doi.org/10.1088/1742-6596/119/6/062011
Apollinari, G., Béjar Alonso, I., Brüning, O. et al: High-luminosity large Hadron Collider (HL-LHC): Technical Design Report V. 0.1. Tech. rep., CERN, (2017) https://doi.org/10.23731/CYRM-2017-004
Arsuaga-Ríos, M., Heikkilä, S.S., Duellmann, D., et al.: Using S3 cloud storage with ROOT and CvmFS. J. Phys. 664(2), 022001 (2015). https://doi.org/10.1088/1742-6596/664/2/022001
Badino, P., Barring, O., Baud, J.P., et al: The Storage Resource Manager Interface Specification (v2.2). (2009) https://sdm.lbl.gov/srm-wg/doc/SRM.v2.2.html
Bevilacqua, G., Bi, H.Y., Hartanto, H.B., et al.: \(\bar{tt}\bar{bb}\) at the LHC: on the size of corrections and b-jet definitions. J. High Energy Phys. 8, 1–37 (2021). https://doi.org/10.1007/JHEP08(2021)008
Bird, I.: Computing for the Large Hadron Collider. Annu. Rev. Nucl. Particle Sci. 61(1), 99–118 (2011). https://doi.org/10.1146/annurev-nucl-102010-130059
Birrittella, M.S., Debbage, M., Huggahalli, R., et al: Intel omni-path architecture: enabling scalable, high performance fabrics. In: 2015 IEEE 23rd Annual Symposium on High-Performance Interconnects, pp 1–9 (2015) https://doi.org/10.1109/HOTI.2015.22
Blomer, J., Canal, P., Naumann, A., et al: Evolution of the ROOT Tree I/O. In: 24th International Conference on Computing in High Energy and Nuclear Physics (CHEP 2019), (2020) https://doi.org/10.1051/epjconf/202024502030
Braam, P.: The Lustre Storage Architecture. (2019) https://arxiv.org/abs/1903.01955
Brun, R., Rademakers, F.: ROOT—an object oriented data analysis framework. Nucl. Instrum. Methods Phys. Res. Sect. A 389(1), 81–86 (1997). https://doi.org/10.1016/S0168-9002(97)00048-X
Calder, B., Wang, J., Ogus, A. et al.: Windows Azure Storage: a highly available cloud storage service with strong consistency. In: Proceedings of the Twenty-Third ACM Symposium on Operating Systems Principles. Association for Computing Machinery, New York, NY, USA, SOSP ’11, pp. 143–157, (2011) https://doi.org/10.1145/2043556.2043571
Carrier, J.: Disrupting high performance storage with intel DC persistent memory & DAOS. In: IXPUG 2019 Annual Conference at CERN. (2019) https://cds.cern.ch/record/2691951
Charbonneau, A., Agarwal, A., Anderson, M., et al.: Data intensive high energy physics analysis in a distributed cloud. J. Phys. 341(012), 003 (2012). https://doi.org/10.1088/1742-6596/341/1/012003
Dai, D., Chen, Y., Kimpe, D., et al.: Provenance-based prediction scheme for object storage system in HPC. In: 2014 14th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing, pp. 550–551, (2014) https://doi.org/10.1109/CCGrid.2014.27
DAOS developers (2022) Caching. https://docs.daos.io/v2.0/user/filesystem/#caching. Accessed 30 July 2022
Din, I.U., Hassan, S., Almogren, A., et al.: PUC: packet update caching for energy efficient IoT-based information-centric networking. Future Gener. Comput. Syst. 111, 634–643 (2020). https://doi.org/10.1016/j.future.2019.11.022
Dorigo, A., Elmer, P., Furano, F., et al.: XROOTD—a highly scalable architecture for data access. WSEAS Trans. Comput. 4, 348–353 (2005)
Elsen, E.: A roadmap for HEP software and computing R &D for the 2020s. Comput. Softw. Big Sci. (2019). https://doi.org/10.1007/s41781-019-0031-6
Hanushevsky, A., Ito, H., Lassnig, M., et al.: Xcache in the atlas distributed computing environment. EPJ Web Conf. 214, 04008 (2019). https://doi.org/10.1051/epjconf/201921404008
ISO Central Secretary (2014) Information technology—Procedures for the operation of object identifier registration authorities—Part 8: Generation of universally unique identifiers (UUIDs) and their use in object identifiers. Standard ISO/IEC 9834-8:2014, International Organization for Standardization, Geneva, CH, https://www.iso.org/standard/62795.html
Jette, M., Dunlap, C., Garlick, J. et al.: Slurm: simple linux utility for resource management. Tech. rep., LLNL, (2002) https://www.osti.gov/biblio/15002962
Kang, G., Kong, D., Wang, L., et al.: OStoreBench: benchmarking distributed object storage systems using real-world application scenarios. In: Wolf, F., Gao, W. (eds.) Benchmarking, Measuring, and Optimizing, pp. 90–105. Springer International Publishing, Cham (2021)
LHCb Collaboration (2017) Matter antimatter differences (b meson decays to three hadrons)—project notebook. http://opendata.cern.ch/record/4902. Accessed 1 Feb 2022
Liang, Z., Lombardi, J., Chaarawi, M., et al.: DAOS: a scale-out high performance storage stack for storage class memory. In: Panda, D.K. (ed.) Supercomputing Frontiers, pp. 40–54. Springer International Publishing, Cham (2020)
Liu, J., Koziol, Q., Butler, G.F. et al.: Evaluation of HPC application I/O on object storage systems. In: 2018 IEEE/ACM 3rd International Workshop on Parallel Data Storage Data Intensive Scalable Computing Systems (PDSW-DISCS), pp. 24–34 (2018) https://doi.org/10.1109/PDSW-DISCS.2018.00005
Lombardi, J.: DAOS: Nextgen Storage Stack for AI, Big Data and Exascale HPC. CERN openlab Technical Workshop. (2021) https://cds.cern.ch/record/2754116
López-Gómez, J., Blomer, J.: Exploring object stores for high-energy physics data storage. EPJ Web Conf. 251(02), 066 (2021). https://doi.org/10.1051/epjconf/202125102066
Matri, P., Alforov, Y., Brandon, A. et al.: Could blobs fuel storage-based convergence between HPC and big data? In: 2017 IEEE International Conference on Cluster Computing (CLUSTER), pp. 81–86, (2017) https://doi.org/10.1109/CLUSTER.2017.63
Mu, J., Soumagne, J., Tang, H. et al.: A transparent server-managed object storage system for HPC. In: 2018 IEEE International Conference on Cluster Computing (CLUSTER), pp. 477–481, (2018) https://doi.org/10.1109/CLUSTER.2018.00063
Padulano, V.E., Cervantes Villanueva, J., Guiraud, E., et al.: Distributed data analysis with ROOT RDataFrame. EPJ Web Conf. 245(03), 009 (2020). https://doi.org/10.1051/epjconf/202024503009
Padulano, V.E., Tejedor Saavedra, E., Alonso-Jordá, P.: Fine-grained data caching approaches to speedup a distributed RDataFrame analysis. EPJ Web Conf. 251(02), 027 (2021). https://doi.org/10.1051/epjconf/202125102027
Panda, D.K., Sur, S.: InfiniBand. Springer, Boston, pp. 927–935. (2011) https://doi.org/10.1007/978-0-387-09766-4_21
Piparo, D., Canal, P., Guiraud, E., et al.: RDataFrame: easy parallel ROOT analysis at 100 threads. EPJ Web Conf. 214(06), 029 (2019). https://doi.org/10.1051/epjconf/201921406029
Plechschmidt, U.: Lustre expands its lead in the Top 100 supercomputers. https://community.hpe.com/t5/Advantage-EX/Lustre-expands-its-lead-in-the-Top-100-supercomputers/ba-p/7141807#.YukqZUhByXJ. Accessed 2 August 2022 (2021)
ROOT team (2021) RNTuple class reference guide. https://root.cern.ch/doc/master/structROOT_1_1Experimental_1_1RNTuple.html. Accessed 1 Feb 2022
ROOT team (2021) TTree class reference guide. https://root.cern.ch/doc/master/classTTree.html. Accessed 1 Feb 2022
Rupprecht, L., Zhang, R., Hildebrand, D.: Big data analytics on object stores : a performance study. In: The International Conference for High Performance Computing, Networking, Storage and Analysis (SC’14) (2014)
Rupprecht, L., Zhang, R., Owen, B. et al.: SwiftAnalytics: optimizing object storage for big data analytics. In: 2017 IEEE International Conference on Cloud Engineering (IC2E), pp. 245–251. https://doi.org/10.1109/IC2E.2017.19 (2017)
Seiz, M., Offenhäuser, P., Andersson, S., et al.: Lustre I/O performance investigations on Hazel Hen: experiments and heuristics. J. Supercomput. 77, 12508–12536 (2021). https://doi.org/10.1007/s11227-021-03730-7
Shin, H., Lee, K., Kwon, H.: A comparative experimental study of distributed storage engines for big spatial data processing using GeoSpark. J. Supercomput. 78, 2556–2579 (2022). https://doi.org/10.1007/s11227-021-03946-7
Soumagne, J., Henderson, J., Chaarawi, M., et al.: Accelerating HDF5 I/O for exascale using DAOS. IEEE Trans. Parallel Distrib. Syst. 33(4), 903–914 (2022). https://doi.org/10.1109/TPDS.2021.3097884
Spiga, D., Ciangottini, D., Tracolli, M., et al.: Smart caching at CMS: applying AI to XCache edge services. EPJ Web Conf. 245, 04024 (2020). https://doi.org/10.1051/epjconf/202024504024
Tang, H., Byna, S., Tessier, F. et al.: Toward scalable and asynchronous object-centric data management for HPC. In: 2018 18th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing (CCGRID), pp. 113–122 (2018) https://doi.org/10.1109/CCGRID.2018.00026
Tannenbaum, T., Wright, D., Miller, K., et al.: Condor—a distributed job scheduler. In: Sterling, T. (ed.) Beowulf Cluster Computing with Linux. MIT Press, New York (2001)
The ATLAS Collaboration, Aad, G., Abat, E., et al.: The ATLAS experiment at the CERN large Hadron Collider. J. Instrum. 3(08), S08003 (2008). https://doi.org/10.1088/1748-0221/3/08/s08003
The LHCb collaboration: angular analysis of the rare decay \(B_s^0 \rightarrow \phi \mu ^+ \mu ^-\). J. High Energy Phys. (2021). https://doi.org/10.1007/JHEP11(2021)043
The LHCb Collaboration, Alves, A.A., Andrade, L.M., et al.: The LHCb Detector at the LHC. JINST 3, S08,005 (2008). https://doi.org/10.1088/1748-0221/3/08/S08005 , also published by CERN Geneva in 2010
Vernik, G., Factor, M., Kolodner, E.K. et al.: Stocator: a high performance object store connector for spark. In: Proceedings of the 10th ACM International Systems and Storage Conference. Association for Computing Machinery, New York, NY, USA, SYSTOR ’17, (2017) https://doi.org/10.1145/3078468.3078496
Vincenzo Eduardo Padulano: Test suite repository. (2021) https://github.com/vepadulano/rdf-rntuple-daos-tests. Accessed 1 Feb 2022
Virgo Cluster: User Manual. (2022) https://hpc.gsi.de/virgo/preface.html. Accessed 2 Aug 2022
Vohra, D.: Apache Parquet, Apress, Berkeley, CA, pp. 325–335. (2016) https://doi.org/10.1007/978-1-4842-2199-0_8
Walker, C.J., Traynor, D.P., Martin, A.J.: Scalable Petascale storage for HEP using Lustre. J. Phys. 396(4), 042063 (2012). https://doi.org/10.1088/1742-6596/396/4/042063
Zhong, J., Huang, R.S., Lee, S.C.: A program for the Bayesian Neural Network in the ROOT framework. Comput. Phys. Commun. 182(12), 2655–2660 (2011). https://doi.org/10.1016/j.cpc.2011.07.019
Acknowledgements
This work benefited from the support of the CERN Strategic R &D Programme on Technologies for Future Experiments [1] and from grant PID2020-113656RB-C22 funded by Ministerio de Ciencia e Innovación MCIN/AEI/10.13039/501100011033. The hardware used to perform the experimental evaluation involving DAOS (HPE Delphi cluster described in Sect. 5.2) was made available thanks to a collaboration agreement with Hewlett-Packard Enterprise (HPE) and Intel. User access to the Virgo cluster at the GSI institute was given for the purpose of running the benchmarks using the Lustre filesystem.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
The following statement is based on CRediT taxonomy, giving contributions of each author in the work: VEP: Conceptualization, Investigation, Writing—original draft, Software, Methodology. ETS: Conceptualization, Supervision, Writing—review and editing, Validation. PA-J: Supervision, Writing—review and editing, Validation. JLG: Conceptualization, Writing—original draft, Software. JB: Conceptualization, Resources
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest in the development of this work.
Informed consent
No informed consent is thus required.
Research involving human rights
This work involved no human subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Padulano, V.E., Tejedor Saavedra, E., Alonso-Jordá, P. et al. A caching mechanism to exploit object store speed in High Energy Physics analysis. Cluster Comput 26, 2757–2772 (2023). https://doi.org/10.1007/s10586-022-03757-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10586-022-03757-2