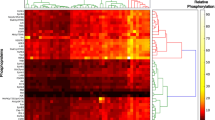
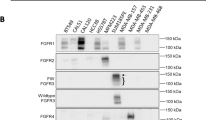
Abstract
Tumor biomarkers assist in the early detection of cancer, act as therapeutic targets for intervention, and function as diagnostic indicators for the evaluation of therapeutic responses. To identify novel human breast cancer biomarkers, we have analyzed the protein content of lipid rafts isolated from a series of human mammary epithelial cell lines with increasing tumorigenic potential. Since lipid rafts function as platforms for protein interaction critical to several biological processes, we hypothesized that the abundance of proteins associated with proliferation, invasion and metastasis would be dysregulated in highly transformed cells. For this purpose, the MCF10A epithelial lineage, which include benign MCF10A cells, premalignant AT and TG3B cells, and malignant CA1a tumor cells, was utilized. Detergent-resistant membranes were isolated from each line and proteins were identified and relatively quantitated using iTRAQ™ reagents and tandem mass spectrometry. 57 proteins were identified, and 1667 peptide identifications, mapping to 49 proteins, contained sufficient information for semi-quantitative analysis. When comparing malignant to benign cells, we observed consistent alterations in groups of proteins, such as a 5.7-fold average decrease in G protein content (n = 5), 2.7-fold decrease in glycosylphosphatidylinositol-linked proteins (n = 7) and 3.3-fold increase in intermediate filaments (n = 9). Several of the identified proteins, including caveolin-1, filamin A, keratins 5, 6 and 17, and vimentin, are bona fide or candidate biomarkers in clinical studies, underscoring the usefulness of the MCF10A series as a model to better understand the biological mechanisms underlying cancer progression.





Similar content being viewed by others
Abbreviations
- DCIS:
-
Ductal carcinoma in situ
- EMT:
-
Epithelial mesenchymal transition
- FDR:
-
False discovery rate
- G protein:
-
Guanine nucleotide-binding proteins
- GPI:
-
Glycosylphosphatidylinositol
- IF:
-
Intermediate filament
- MudPIT:
-
Multidimensional protein identification technology
References
Hanzal-Bayer MF, Hancock JF (2007) Lipid rafts and membrane traffic. FEBS Lett 581:2098–2104
Lingwood D, Simons K (2010) Lipid rafts as a membrane-organizing principle. Science 327:46–50
Patra SK (2008) Dissecting lipid raft facilitated cell signaling pathways in cancer. Biochim Biophys Acta 1785:182–206
Lajoie P, Goetz JG, Dennis JW et al (2009) Lattices, rafts and scaffolds: domain regulation of receptor signaling at the plasma membrane. J Cell Biol 185(3):381–385
Soule HD, Maloney TM, Wolman SR et al (1990) Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res 50:6075–6086
Russo J, Tait L, Russo IH (1991) Morphological expression of cell transformation induced c-Ha-ras oncogene in human breast epithelial cells. J Cell Sci 99:453–463
Dawson PJ, Wolman SR, Tait L et al (1996) MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol 148(1):313–319
Santner SJ, Dawson PJ, Tait L et al (2001) Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat 65:101–110
Ostrom RS, Liu X (2007) Detergent and detergent-free methods to define lipid rafts and cavaeolae. Methods Mol Biol 400:459–468
Griffin TJ, Xie H, Bandhakavi S et al (2007) iTRAQ reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. J Proteome Res 6:4200–4209
Feng J, Xie H, Meany DL et al (2008) Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J Gerontol 63A(11):1137–1152
Caruso JA, Reiners JJ Jr (2006) Proteolysis of HIP during apoptosis occurs within a region similar to the BID loop. Apoptosis 11(11):1877–1885
Caruso JA, Mathieu PA, Reiners JJ Jr (2005) Sphingomyelins suppress the targeted disruption of lysosomes/endosomes by the photosensitizer NPe6 during photodynamic therapy. Biochem J 392(2):325–334
Feng Y, Walsh CA (2004) The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol 6:1034–1038
Alper O, Stetler-Stevenson WG, Harris LN et al (2009) Novel anti-filamin-A antibody detects a secreted variant of filamin-A in plasma from patients with breast carcinoma and high-grade astrocytoma. Cancer Sci 100(9):1748–1756
Parton RG (1994) Ultrastructural localization of gangliosides; GM1 is concentrated in caveolae. J Histochem Cytochem 42(2):155–166
Mishra S, Ande SR, Grégoire Nyomba LG (2010) The role of prohibitin in cell signaling. FEBS J 277(19):3937–3946
Staubach S, Razawi H, Hanisch FG (2009) Proteomics of MUC1-containing lipid rafts from plasma membranes and exosomes of human breast carcinoma cells MCF-7. Proteomics 9(10):2820–2835
Kim KB, Lee JW, Lee CS et al (2006) Oxidation-reduction respiratory chains and ATP synthase complex are localized in detergent-resistant lipid rafts. Proteomics 6(8):2444–2453
Ponce J, Brea D, Carrascal M et al (2010) The effect of simvastatin on the proteome of detergent-resistant membrane domains: decreases of specific proteins previously related to cytoskeletal regulation, calcium homeostasis and cell fate. Proteomics 10(10):1954–1965
Ande SR, Mishra S (2010) Palmitoylation of prohibitin at cysteine 69 facilitates its membrane translocation and interaction with Eps 15 homology domain protein 2 (EHD2). Biochem Cell Biol 88(3):553–558
Coates PJ, Nenutil R, McGregor RA et al (2001) Mammalian prohibitin proteins respond to mitochondrial stress and decrease during cellular senescence. Exp Cell Res 265(2):262–273
Stratford AL, Reipas K, Maxwell C et al (2010) Targeting tumour-initiating cells to improve the cure rates for triple-negative breast cancer. Expert Rev Mol Med 12:e22
Alvarez RH, Valero V, Hortobagyi GN (2010) Emerging targeted therapies for breast cancer. J Clin Oncol 28(20):3366–3379
Baruthio F, Quadroni M, Ruegg C et al (2008) Proteomic analysis of membrane rafts of melanoma cells identifies protein patterns characteristic of the tumor progression stage. Proteomics 8:4733–4747
Patel HH, Murray F, Insel PA (2008) G-protein-couples receptor-signaling components in membrane raft and caveolae microdomains. Handb Exp Pharmacol 186:167–184
Sharom FJ, Lehto MT (2002) Glycosylphosphatidylinositol-anchored proteins: structure, function, and cleavage by phosphatidylinositol-specific phospholipase C. Biochem Cell Biol 80:535–549
Mann KJ, Hepworth MR, Raikwar NS (2004) Effect of glycosylphosphatidylinositol (GPI)-phospholipase D overexpression on GPI metabolism. Biochem J 378:641–648
He X, Hannocks MJ, Hampson I et al (2002) GPI-specific phospholipase D mRNA expression in tumor cells of different malignancy. Clin Exp Metastasis 9(4):291–299
Birchmeier C, Birchmeier W, Brand-Saberi B (1996) Epithelial mesenchymal transitions in cancer progression. Acta Anat 156:217–226
Kokkinos MI, Wafai R, Wong MK et al (2007) Vimentin and epithelial-mesenchymal transition in human breast cancer—observations in vitro and in vivo. Cells Tissues Organs 185(1–3):191–203
Thomas PA, Kirschmann DA, Cerhan JR (1999) Association between keratin and vimentin expression, malignant phenotype, and survival in postmenopausal breast cancer patients. Clin Cancer Res 5:2698–2703
Chen MH, Yip GW, Tse GM et al (2008) Expression of basal keratins and vimentin in breast cancers of young women correlates with adverse pathologic parameters. Mod Pathol 21:1183–1191
Dabbs DJ, Chivukula M, Carter G (2006) Basal phenotype of ductal carcinoma in situ: recognition and immunohistologic profile. Mod Pathol 19:1506–1511
Kuroda N, Ohara M, Inoue K et al (2009) The majority of triple-negative breast cancer may correspond to basal-like carcinoma, but triple-negative breast cancer in not identical to basal-like carcinoma. Med Mol Morphol 42:128–131
van de Rijn M, Perou CM, Tibshirani R et al (2002) Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol 161(6):1991–1996
Hendrix MJC, Seftor EA, Seftor REB et al (1997) Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am J Pathol 150:483–495
McInroy L, Maatta A (2007) Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem Biophys Res Commun 360:109–114
Choong LY, Lim S, Chong PK et al (2010) Proteome-wide profiling of the MCF10AT breast cancer progression model. PLoS One 5(6):e11030
Lee SW, Reimer CL, Oh P et al (1998) Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells. Oncogene 16:1391–1397
Koleske AJ, Baltimore D, Lisanti MP (1995) Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci USA 92:1381–1385
Mercier I, Casimiro MC, Wang C et al (2008) Human breast cancer-associated fibroblasts (CAFs) show caveolin-1 downregulation and RB tumor suppressor functional inactivation: implications for the response to hormonal therapy. Cancer Biol Ther 7:1212–1225
Wua P, Wang X, Lib F et al (2008) Growth suppression of MCF-7 cancer cell-derived xenografts in nude mice by caveolin-1. Biochem Biophys Res Commun 376(1):215–220
Fiucci G, Ravid D, Reich R et al (2002) Caveolin-1 inhibits anchorage-independent growth, anoikis and invasiveness in MCF-7 human breast cancer cells. Oncogene 21(15):2365–2375
Williams TM, Medina F, Badano I et al (2004) Caveolin-1 gene disruption promotes mammary tumorigenesis and dramatically enhances lung metastasis in vivo. Role of Cav-1 in cell invasiveness and matrix metalloproteinase (MMP-2/9) secretion. J Biol Chem 279(49):51630–51646
Perrone G, Altomare V, Zagami M et al (2009) Caveolin-1 expression in human breast lobular cancer progression. Mod Pathol 22:71–78
Weigelt B, Geyer FC, Natrajan R, Lopez-Garcia MA, Ahmad AS, Savage K, Kreike B, Reis-Filho JS (2010) The molecular underpinning of lobular histological growth pattern: a genome-wide transcriptomic analysis of invasive lobular carcinomas and grade- and molecular subtype-matched invasive ductal carcinomas of no special type. J Pathol 220(1):45–57
Sloan EK, Ciocca DR, Pouliot N et al (2009) Stromal cell expression of caveolin-1 predicts outcome in breast cancer. Am J Pathol 174(6):2035–2043
Witkiewicz AK, Dasgupta A, Nguyen KH et al (2009) Stromal caveolin-1 levels predict early DCIS progression to invasive breast cancer. Cancer Biol Ther 8(11):1071–1079
Witkiewicz AK, Dasgupta A, Sammons S et al (2010) Loss of stromal caveolin-1 expression predicts poor clinical outcome in triple negative and basal-like breast cancers. Cancer Biol Ther 10(2):135–143
Yang G, Truong LD, Timme TL et al (1998) Elevated expression of caveolin is associated with prostate and breast cancer. Clin Cancer Res 4(8):1873–1880
Pinilla SM, Honrado E, Hardisson D et al (2006) Caveolin-1 expression is associated with a basal-like phenotype in sporadic and hereditary breast cancer. Breast Cancer Res Treat 99(1):85–90
Savage K, Lambros MB, Robertson D et al (2007) Caveolin 1 is overexpressed and amplified in a subset of basal-like and metaplastic breast carcinomas: a morphologic, ultrastructural, immunohistochemical, and in situ hybridization analysis. Clin Cancer Res 13(1):90–101
Elsheikh SE, Green AR, Rakha EA et al (2008) Caveolin 1 and caveolin 2 are associated with breast cancer basal-like and triple-negative immunophenotype. Br J Cancer 99(2):327–334
Mammoto A, Huang S, Ingber DE (2007) Filamin links cell shape and cytoskeletal structure to Rho regulation by controlling accumulation of p190RhoGAP in lipid rafts. J Cell Sci 120:456–467
Tavano R, Contento RL, Baranda SJ et al (2006) CD28 interaction with filamin-A controls lipid raft accumulation at the T-cell immunological synapse. Nat Cell Biol 8(11):1270–1276
Ravid D, Chuderland D, Landsman L et al (2008) Filamin A is a novel caveolin-1-dependent target in IGF-I-stimulated cancer cell migration. Exp Cell Res 314(15):2762–2773
Xu Y, Bismar TA, Su J et al (2010) Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J Exp Med 207(11):2421–2437
Acknowledgments
This research has been supported by the Proteomics Facility Core of the Institute of Environmental Health Sciences at Wayne State University, which is supported by National Institute of Environmental Health Sciences grant P30-ES006639.
Conflict of interest
The authors declare no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Subcellular localization of prohibitin-2 in the MCF10A series of human breast cell lines. Cells were fixed and stained according to procedures described in “Methods”. Two independent photographs are shown per cell line. Exposure times and contrast adjustments remained constant throughout the experiment. Prohibitin-2 staining is shown in green, and nuclei are stained blue. This experiment was repeated with similar results. (PDF 104 kb)
Rights and permissions
About this article
Cite this article
Caruso, J.A., Stemmer, P.M. Proteomic profiling of lipid rafts in a human breast cancer model of tumorigenic progression. Clin Exp Metastasis 28, 529–540 (2011). https://doi.org/10.1007/s10585-011-9389-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-011-9389-5