Abstract
A fundamental component of user-level social media language based clinical depression modelling is depression symptoms detection (DSD). Unfortunately, there does not exist any DSD dataset that reflects both the clinical insights and the distribution of depression symptoms from the samples of self-disclosed depressed population. In our work, we describe a semi-supervised learning (SSL) framework which uses an initial supervised learning model that leverages (1) a state-of-the-art large mental health forum text pre-trained language model further fine-tuned on a clinician annotated DSD dataset, (2) a Zero-Shot learning model for DSD, and couples them together to harvest depression symptoms related samples from our large self-curated depressive tweets repository (DTR). Our clinician annotated dataset is the largest of its kind. Furthermore, DTR is created from the samples of tweets in self-disclosed depressed users Twitter timeline from two datasets, including one of the largest benchmark datasets for user-level depression detection from Twitter. This further helps preserve the depression symptoms distribution of self-disclosed tweets. Subsequently, we iteratively retrain our initial DSD model with the harvested data. We discuss the stopping criteria and limitations of this SSL process, and elaborate the underlying constructs which play a vital role in the overall SSL process. We show that we can produce a final dataset which is the largest of its kind. Furthermore, a DSD and a Depression Post Detection model trained on it achieves significantly better accuracy than their initial version.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
1 Introduction
According to Boyd et al. (1982), in developed countries, around 75% of all psychiatric admissions are young adults with depression. The fourth leading cause of death in young adults is suicide, which is closely related to untreated depression (World Health Organization, 2023). Moreover, traditional survey-based depression screening may be in-effective due to the cognitive bias of the patients who may not be truthful in revealing their depression condition. So there is a huge need for an effective, inexpensive and almost real time intervention for depression in this high risk population. Interestingly, among young adults, social media is very popular where they share their day to day activities and the availability of social media services is growing exponentially year by year (O’Keeffe & Clarke-Pearson, 2011). Moreover, according to the research (Gowen et al., 2012; Naslund et al., 2014, 2016), it has been found that depressed people who are otherwise socially aloof, show increased use of social media platforms to share their daily struggles, connect with others who might have experienced the same and seek help. So, in this research we focus on identifying depression symptoms from a user’s social media posts as one of the strategies for early identification of depression. Earlier research confirms that signs of depression can be identified in the language used in social media posts (Coppersmith et al., 2015; De Choudhury & De, 2014; De Choudhury et al., 2013; Losada & Crestani, 2016; Reece et al., 2017; Rude et al., 2004; Seabrook et al., 2018; Shen et al., 2017; Trotzek et al., 2018; Yadav et al., 2020; Yazdavar et al., 2017). Based on this background, linguistic features, such as n-grams, psycholinguistic and sentiment lexicons, word and sentence embeddings extracted from the social media posts can be very useful for detecting depression, especially when compared to other social media related features which are not language specific, such as social network structure of depressed users and their posting behavior. In addition, the majority of this background research focused on public social media data, i.e., Twitter and Reddit mental health forums for user-level depression detection, because of the relative ease of accessing such datasets (unlike Facebook and other social media which have strict privacy policies). All this background placed emphasis on signs of depression detection, however, they lacked the inclusion of clinical depression modelling; such requires extensive effort in building a depression symptoms detection model (Sect. 4.2). Some of the earlier research (Ma et al., 2017; Mowery et al., 2016; Safa et al., 2022; Tlelo-Coyotecatl et al., 2022; Yazdavar et al., 2017; Yadav et al., 2020) has focused on depression symptoms detection but they do not attempt to create a clinician-annotated dataset, and later use existing state-of-the-art language models to expand it. All the previous research does not attempt to curate the possible depression candidate dataset from self-disclosed depressed users’ timelines. Therefore the main motivation of this work arises from the following:
-
1.
Clinician-annotated dataset creation from depressed users tweets: Through leveraging our existing datasets from self disclosed depressed users and trained Depression Post Detection (DPD) model (which is a binary model for detecting signs of depression), we want to curate a clinician-annotated dataset for depression symptoms. This is a more “in-situ” approach for harvesting depression symptoms posts compared to crawled tweets for depression symptoms using depression symptoms keywords, as done in most of the earlier literature (Mowery et al., 2016, 2017). We call it in-situ because this approach respects the natural distribution of depression symptoms samples found in the self-disclosed depressed users’ timelines. Although Yadav et al. (2020) collected samples in-situ as well, our clinician-annotated dataset is much bigger and annotation is more rigorous (Sect. 5.1).
-
2.
Gather more data that reflects clinical insight: Starting from the small dataset found at (1) and a DSD model trained on that, we want to iteratively harvest more data and retrain our model for our depression symptoms modelling or DSD task.
Our dataset made of both clinician annotated and harvested tweets with signs of depression symptoms is the largest of its kind, to the best of our knowledge.
2 Methodology
To achieve the goals mentioned earlier, we divide our depression symptoms modelling into two parts: (1) Clinician annotated dataset curation: here we first propose a process to create our annotation candidate dataset from our existing depressive tweets from self-disclosed depressed Twitter users. We later annotate this dataset with the help of a clinician amongst others, that helps us achieve our first goal (Sect. 3) and (2) Semi-supervised Learning (SSL): we then describe how we leverage that dataset to learn our first sets of DPD and DSD models and eventually make them robust through iterative data harvesting and retraining or SSL (McClosky et al., 2006) (Sect. 4).
3 Datasets
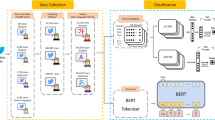
We create Depression-Candidate-Tweets dataset from the timeline of depressed users in IJCAI-2017 (Shen et al., 2017) who disclosed their depression condition through a self-disclosure statement, such as: "I (am / was / have) been diagnosed with depression" and UOttawa (Jamil et al., 2017) datasets where the users were verified by annotators about their ongoing depression episodes. Later, we further filter it with a DPD model (discussed in Sect. 3.1) for depressive tweets and create the depressive tweets repository (DTR) which is used in our SSL process to harvest in-situ tweets for depression symptoms. We also separate a portion of the DTR for clinician annotation for depression symptoms (Fig. 3).
3.1 Clinician annotated dataset curation
In the overall DSD framework, depicted in Fig. 1, we are ultimately interested in creating a robust DPD and a DSD model which are initially trained on human annotated samples, called “DPD-Human” model and “DSD-Clinician” model as depicted in Fig. 2. The suffixes with these model names, such as “Human,” indicates that this model leverages the annotated samples from both non-clinicians and clinicians; “Clinician” indicates that this model leverages the samples for which the clinician’s annotation is taken as more important (more explanation is provided later in Sect. 3.4). At the beginning of this process, we have only a small human annotated dataset for depression symptoms augmented with depression posts from external organizations (i.e. D2S (Yadav et al., 2020) and DPD-Vioules (Vioulès et al., 2018) datasets), no clinician annotated depression symptoms samples, and a large dataset from self-disclosed depressed users (i.e IJCAI-2017 dataset). We take the following steps to create our first set of clinician annotated depression symptoms dataset and DTR which we will use later for our SSL.
-
1.
We start the process with the help of a DPD model, which we call DPD Majority Voting model (DPD-MV). It consists of a group of DPD models (Farruque et al., 2019), where each model leverages pre-trained word embedding (both augmented (ATE) and depression specific (DSE)) and sentence embedding (USE), further trained on a small set of human annotated depressive tweets and a Zero-Shot Learning (ZSL) model (USE-SE-SSToT). This ZSL model helps determine the semantic similarity between a tweet and all the possible depression symptoms descriptors and returns the top-k corresponding labels. It also provides a score for each label, based on cosine distance. More details are provided in a previous paper (Farruque et al., 2021). Subsequently, the DPD-MV model takes the majority voting of these models for detecting depressive tweets.
-
2.
We then apply DPD-MV on the sets of tweets collected from depressed users’ timelines (or Depression-Candidate-Tweets, (Fig. 3) to filter control tweets. The resultant samples, after applying DPD-MV is referred to as Depression Tweet Repository or DTR. We later separate a portion of this dataset, e.g., 1500 depressive tweets for human annotation which we call DSD-Clinician-Tweets dataset. Details of the annotation process are described in Sect. 3.4.
-
3.
We train our first DSD model using this dataset, then use this model to harvest more samples from DTR. An outline of the DTR and DSD-Clinician-Tweets curation process is provided in Fig. 3. We describe the details of this process in Sect. 4.2, but describe each of its building blocks in the next sections. In Table 1 we provide relevant datasets description.
3.2 Annotation task description
Our annotation task consists of labelling a tweet for either (1) one or more of 10 symptoms of depression (See next section), (2) No Evidence of Depression (NoED), (3) Evidence of Depression (ED) or (4) Gibberish. We have 10 labels instead of the traditional nine depression symptoms labels because we separate the symptom “Agitation / Retardation” into two categories so that our model can separately learn and distinguish these labels, unlike previous research (Yadav et al., 2020). NoED indicates the absence of any depression symptoms expressed in a tweet. ED indicates multiple symptoms of depression expressed in a tweet in a way so that it’s hard to specifically pinpoint these combined depression symptoms in that tweet. Gibberish is a tweet less than three words long and, due to the result of crawling or data pre-processing, the tweet is not complete and it’s hard to infer any meaningful context.
3.3 Annotation guideline creation
To create the annotation guideline for the task, we analyze the textual descriptions of depression symptoms from most of the major depression rating scales, such as, PHQ-9, CES-D, BDI, MADRS and HAM-D (The classification of depression, 2010). We also use DSM-5 as our reference for symptom descriptions. Based on these descriptions of the symptoms from these resources and several meetings with our clinicians, we consolidate some of the most confusing samples of tweets from DTR and map them to one or more of those depression symptoms. We then create an annotation guideline with a clear description of the clinical symptoms of depression that an annotator should look for in the tweets followed by relevant tweet examples for them including the confusing ones previously noted. We then separate a portion of 1500 samples from our DTR and provide it to the annotators along with our annotation guideline. During the annotation, we randomly assign a set of tweets multiple times to calculate test-retest reliability scores. We find annotators annotate the tweets consistently with the same annotation with 83% reliability based on the test-retest reliability score. Our detailed guideline description is provided in Appendix 3.
3.4 Depression symptoms annotation process
We provide a portion of 1500 tweets from DTR for depression symptoms annotation by four annotators.
Among these annotators two have a clinical understanding of depression: one is a practicing clinician and the other one has a Ph.D. in Psychiatry.
Our annotation process is based on the clinical understanding of depression as outlined in our guidelines. We take majority voting to assign a label for the tweet. In the absence of a majority, we assign a label based on the clinician’s judgment, if present, otherwise, we do not assign a label to that tweet. We call this scheme Majority Voting with Clinician Preference (MVCP). Table 2 reports the average Cohen’s kappa scores for each label and Annotator-Annotator, Annotator-MVCP, and All pairs (i.e. avg. on both of the previous schemes). Through out the paper by kappa score, we mean Cohen’s kappa score.
We observe fair to moderate kappa agreement score (0.38–0.53) among our annotators for all the labels. We also find, “Suicidal thoughts” and “Change in Sleep Patterns” are the labels for which inter-annotator agreement is the highest and agreement between each annotator and MVCP is substantial for the same. Among the annotators the order of the labels based on descending order of agreement score is as follows: Suicidal thoughts, Change in Sleep Patterns, Feelings of Worthlessness, Indecisiveness, Anhedonia, Retardation, Weight change, NoED, Fatigue, Low mood, Gibberish, Agitation and ED. However, with MVCP, we find moderate to substantial agreement (0.56–0.66). For all labels and annotators, we find a global inter-annotator agreement score (Krippendorff’s alpha) of 0.3064.
3.5 Distribution analysis of the depression symptoms data
In this section, we provide symptom distribution analysis for D2S and DSD-Clinician-Tweets datasets. DSD-Clinician-Tweets dataset contains 1500 tweets. We then create a clean subset of this dataset which holds clinicians’ annotations and only tweets with depression symptoms, which we call DSD-Clinician-Tweets-Original (further detail is in Sect. 4.2.1). For D2S, we have 1584 tweets with different depression symptom labels. In Fig. 4, the top 3 most populated labels for the DSD dataset are "Agitation", "Feeling of Worthlessness", and "Low Mood". However, for the D2S dataset, "Suicidal Thought" is the most populated label followed by “Feelings of Worthlessness” and “Low Mood”, just like DSD. We use the D2S dataset because D2S crawled tweets from self-reported depressed users’ timeline. Although they did not confirm whether these users have also disclosed their depression diagnosis, they mention that they analyze their profile to ensure that these users are going through depression. Since their annotation process is not as rigorous as ours, i.e., they did not develop an annotation guideline as described in the earlier section and their dataset may not contain all self-disclosed depressed users, we had to further filter those tweets before we could use them. So we use DSD-Clinician-Original-Tweets for training our very first model in the SSL process, and later use that to re-label D2S samples.
In Sect. 4.2.6, we report the distribution on harvested data and another approach for increasing sample size for least populated labels.
4 Experimental setup and evaluation
Our experimental setup consists of iterative data harvesting and re-training of a DSD and a DPD model (Sect. 4.2), followed by observing their accuracy increase over each iteration coupled with incremental initial dataset size increase.
We report the results separately for each of the steps of SSL in the next sections. For the DSD task, which is a multi-class multi-label problem, we report Macro and Weighted-Averaged Precision, Recall, and F1 along with label-wise Precision, Recall, and F1 scores as our accuracy scores. Macro-F1 is an average F1 score for all the labels, whereas weighted F1-score is a measure that assigns more weight to the labels for which we have the most samples. For the DPD task, which is a binary classification problem, we report Macro-Averaged Precision, Recall, and F1 scores as our accuracy scores.
From our clinician-annotated dataset, we separate a subset of depression symptoms stratified samples as a test-set for the DSD task. For the DPD task, we separate a 10% portion from the DPD-Human train-set as a test set. After each step of the SSL process, we report the accuracy scores to evaluate the efficacy of that step based on the DSD and DPD models’ performance on these test-sets respectively (Tables 3 and 4).
4.1 Data preprocessing
We perform the following preprocessing steps for all our Twitter datasets, we use NLTKFootnote 1 for tokenizing our tweets and also EkphrasisFootnote 2 for normalizing tweets.
-
1.
Lowercase each word.
-
2.
Remove one character words and digits.
-
3.
De-contract contracted words in a tweet. For example, “I’ve” is made “I have”.
-
4.
Elongated words are converted to their original form. For example, “Looong” is turned into “Long”.
-
5.
Remove tweets with self-disclosure, i.e. any tweet containing the word “diagnosed” or “diagnosis” is removed.
-
6.
Remove all punctuations except period, comma, question mark, and exclamation.
-
7.
Remove URLs.
-
8.
Remove non-ASCII characters from words.
-
9.
Remove hashtags.
-
10.
Remove emojis.
4.2 Semi-supervised learning (SSL) framework
In our SSL framework, we iteratively perform data harvesting and retraining of our DSD model, which is a multi-label text classifier utilizing pre-trained Mental-BERT,Footnote 3 technical details of this model (i.e., the training hyper-parameters) are provided in Appendix 2. We find Mental-BERT-based DSD performs significantly better in terms of Macro-F1 and Weighted-F1 scores compared to base BERT-only models in the DSD task (Tables 5 and 6). In this section, we provide our step-by-step SSL process description, datasets utilized at each step, and the resulting models and/or datasets.
All our steps are depicted in points 11–25 in Fig. 5 and described further below.
Detailed SSL framework. Here, we show the interaction among our datasets and models. Datasets are shown as cylinders, models are shown as rectangles. An arrow from a dataset to another dataset represents data subset creation; an arrow to another model means the provision of training data for that model; and an arrow from a model to a dataset means the use of that model to harvest samples from the dataset. All the arrow heads are marked, so that these can be easily referred while describing a particular scenario in the SSL framework
4.2.1 Step 1: creating first DSD model
In this step, we focus on the creation of a training dataset and a test dataset selected from our clinician-annotated samples. This dataset consists of tweets carrying at least one of the 10 depression symptoms. We use this training dataset to create our first DSD model, called DSD-Clinician-1. To do so, we follow the steps stated below.
-
1.
We first remove all the tweets with labels “Gibberish,” “Evidence of Depression” (ED) and “No Evidence of Depression” (NoED) from a subset of DSD-Clinician-Tweets after applying MVCP. We call this dataset DSD-Clinician-Tweets-Original. Details of ED, NoED, and Gibberish are provided in Table 3.
-
2.
We save the tweets labelled as “Evidence of Depression,” which we call DSD-Clinician-ED-Tweets, (Arrow 8 in Fig. 5). We later use those to harvest depression symptoms-related tweets.
-
3.
Next, we separate 70% of the tweets from DSD-Clinician-Tweets-Original dataset and create DSD-Clinician-Tweets-Original-Train dataset for training our first version of DSD model, called DSD-Clinician-1 and the rest 30% of the tweets are used as an SSL evaluation set, also called, DSD-Clinician-Tweets-Original-Test, (Arrows 5 and 7 in the Fig. 5). We will use this evaluation set all through our SSL process to measure the performance of SSL, i.e., whether it helps increase accuracy for DSD task or not. We report the datasets created in this step in Table 3, models in Table 4, and accuracy scores for each label and their average in Table 6. We also report the accuracy for the DPD-Human model in this step in Table 7.
Table 7 DPD-Human model accuracy in step 1
4.2.2 Step 2: harvesting tweets using DSD-Clinician-1
In this step, we use DSD-Clinician-1 model created in the previous step to harvest tweets that carry signs of depression symptoms from a set of tweets filtered for carrying signs of depression only by DPD-Human model from DTR, we call this dataset DSD-Harvest-Candidate-Tweets (Arrows 10 and 12 in Fig. 5). Our DPD-Human model is trained on all available human annotated datasets, i.e., DSD-Clinician-Tweets-Original, D2S, and an equal number of control tweets from DTR (Arrows 6 and 9 in Fig. 5 and more dataset details in Table 4). We use this model to leverage human insights to further filter DTR. In this step, we create two more datasets from DSD-Harvest-Candidate-Tweets, (1) Harvested-DSD-Tweets: This dataset contains the tweet samples for which the model is confident, i.e., it detects one of the 10 depression symptoms and (2) Harvested-DSD-Tweets-Less-Confident: This dataset contains the tweet samples for which the model has no confident predictions or it does not predict any depression symptoms for harvested dataset (Table 8).
4.2.3 Step 3: harvesting tweets using best ZSL Model
In this step, we use a ZSL model (USE-SE-SSToT) described in Farruque et al. (2021) to harvest tweets carrying signs of depression symptoms from the DSD-Harvest-Candidate-Tweets. We chose this model because it has reasonable accuracy in the DSD task and it is fast. We also set a threshold while finding semantic similarity between the tweet and the label descriptor to be more on the conservative side so that we reduce the number of false positive tweets. We find that a threshold < 1 is a reasonable choice because cosine-distance < 1 indicates higher semantic similarity. In this step, we create two datasets: (1) Only-ZSL-Pred-on-Harvested-DSD-Tweets (step: 3a): This dataset is only ZSL predictions on DSD-Harvest-Candidate-Tweets. (2) ZSL-and-Harvested-DSD-Tweets (step: 3b): This dataset is a combination of ZSL predictions and DSD-Clinician-1 predictions on DSD-Harvest-Candidate-Tweets. We follow steps: 3a and 3b to compare whether datasets produced through these steps help in accuracy gain after using them to retrain DSD-Clinician-1.
Compared to step 1 (Table 6), we achieve 4% gain in Macro-F1 and 5% gain in Weighted-F1 using the combined dataset in step: 3b (Table 10). We achieve 1% gain in both the measures using Harvested-DSD-Tweets only in step: 2 (Table 9). With ZSL only in step: 3a (Table 11), we lose 3% in Macro-F1 and 15% in Weighted-F1. We also provide our produced datasets description in Table 12.
4.2.4 Step 4: creating a second DSD Model:
From the previous experiments, we now create our second DSD model by retraining it with DSD-Clinician-Tweets-Original-Train and ZSL-and-Harvested-DSD-Tweets. This results in our second DSD (DSD-Clinician-2) model (Table 13).
4.2.5 Step 5: creating final DSD model
In this final step, we do the following:
-
1.
We create a combined dataset from D2S and DSD-Clinician-ED-Tweets and we call this combined dataset DSD-Less-Confident-Tweets dataset (Arrows 15, 16, 17, 20 in Fig. 5). D2S tweets are used here because the dataset was annotated externally with a weak clinical annotation guideline. We use our model to further filter this dataset.
-
2.
We use DSD-Clinician-2 model and ZSL to harvest depression symptoms tweets from DSD-Less-Confident-Tweets, we call this dataset Harvested-DSD-from-Less-Confident-Tweets. Finally, with this harvested data and the datasets used to train DSD-Clinician-2 model, we create our final dataset called Final-DSD-Clinician and by training with it, we learn our final DSD model called, Final-DSD-Clinician. We also retrain our DPD-Human model to create Final-DPD-Human model. Datasets, models, and the relevant statistics are reported in Tables 14, 15, 16 and 17. We reported the symptoms distribution for our DSD-Clinician-Tweets-Original-Train dataset earlier, and here report depression symptoms distribution in our SSL model harvested datasets (ZSL-and-Harvested-DSD-Tweets + Harvested-DSD-from-Less-Confident-Tweets) only (Fig. 6). We see that the sample size for all the labels generally increases and reflects almost the same distribution as our DSD-Clinician-Tweets-Original-Train dataset. Interestingly, data harvesting increases the sample size of “Feelings of Worthlessness” and “Suicidal thoughts” while still maintaining the distribution of our original clinician annotated dataset (DSD-Clinician-Tweets-Original-Train) (Fig. 6). We also report the top-10 bi-grams for each of the symptoms for our Final-DSD-Clinician-Tweets dataset in Table 18. We see that top bi-grams convey the concepts of each symptoms.
4.2.6 Step 6: combating low accuracy for less populated labels
Here we attempt to combat the low accuracy for the labels that have a very small sample size. In these cases, we analyze the co-occurrence of those labels with other labels through an associative rule mining (Apriori) algorithm (Agrawal et al., 1994). Our idea is to use significant co-occurring labels and artificially predict one label if the other occurs. For that, we analyze a small human-annotated train dataset (DSD-Clinician-Tweets-Original-Train). However, since the support and confidence for association rules are not significant due to the small sample size, we consider all the “strong” rules with non-zero support and confidence scores for those labels. The rules we consider have the form: (strong-label \(\rightarrow\) weak-label), where the weak label (such as Anhedonia, Fatigue, Indecisiveness, and Retardation) means the labels for which our model achieves either 0 F1 score or very low recall, i.e., less or equal to chance level. These are the candidate labels for which we would like to have increased accuracy. On the other hand, strong labels are those for which we have at least a good recall, i.e., beyond chance level. By emphasizing high recall, we intend to prevent a depression symptom from being undetected by our model. All the extracted strong rules are provided in Appendix 1. When we compare the sample distribution for Apriori-based harvested data and plain harvested data, we see for the least populated class we have more samples (Fig. 7). This makes the classification task more sensitive towards weak labels. However, with this method, we do not achieve a better Macro-F1 score compared to our Final-DSD-Clinician model (Table 19).
4.2.7 Stopping criteria for SSL
The following two observations lead us to stop the SSL:
-
1.
Our DTR consists of a total 6077 samples and we have finally harvested 4567 samples, so for \((6077-4567)=1510\) samples neither ZSL nor any version of DSD models have any predictions. We exhausted all our depression candidate tweets from all sources we have, therefore, we do not have any more depression symptoms candidate tweets for moving on with SSL.
-
2.
We have another very noisy dataset, called IJCAI-2017-Unlabelled (Shen et al., 2017), where we have tweets from possible depressed users, i.e., their self-disclosure contains the stem “depress” but it is not verified whether they are genuine self-disclosures of depression. Using our Final-DSD-Clinician model we harvest \(\approx 22K\) depression symptoms tweets from \(\approx 0.4M\) depression candidate tweets identified by the Final-DPD-Human model from that dataset. We then retrain the Final-DSD-Clinician model on all the samples previously we harvested combined with the newly harvested \(\approx 22K\) tweets, which results in a total of \(\approx 26k\) tweets (\(\approx 6\) times larger than the samples DSD-Final-model was trained on). However, we did not see any significant accuracy increase, so we did not proceed (Table 20).
Table 20 DSD-Clinician model trained on IJCAI-2017-Unlabelled and all the harvested dataset
5 Results analysis
Here we analyse the efficacy of our SSL frameworks in three dimensions, as follows:
5.1 Dataset size increase
Through the data harvesting process, we can increase our initial clinician annotated 377 samples to 4567 samples, which is 12 times bigger than our initial dataset. In addition, we have access to an external organization-collected dataset (i.e., D2S), for which we could access around \(\approx 1800\) samples. Our final dataset is more than double the size of that dataset.
5.2 Accuracy improvement
Our Final-DSD-Clinician model has Macro-F1 score of 45% which is 14% more than that of our initial model and Weighted-F1 score increased by 5% from 51% to 56% (Table 21). The substantial gain in the Macro-F1 score indicates the efficacy of our data harvesting in increasing F1 scores for all the labels. We also find that the combination of DSD-Clinician-1 and ZSL models in step 3a helps achieve more accuracy than individually; specifically, using only ZSL-harvested data for training is not ideal. Weighted-F1 has slow growth and does not increase after Step 3b. We also find that the combined harvesting process on D2S samples helped us achieve further accuracy in a few classes for which D2S had more samples, such as “Fatigue,” “Weight Change” and “Suicidal Thoughts.”
5.3 Linguistic components distribution
In Table 18, we see that our harvested dataset contains important clues about depression symptoms. Interestingly, there are some bi-grams, such as, “feel like” occur in most of the labels; this signifies the frequent usage of that bi-gram in various language-based expressions of depression symptoms. This also shows a pattern of how people describe their depression.
5.4 Sample distribution
Compared with the original clinician annotated dataset distribution (Fig. 6), we see similar trends in our harvested dataset, i.e., in Final-DSD-Clinician-Tweets. However, instead of “Agitation” we have some more samples on “Feeling of Worthlessness,” although those are not surpassed by “Suicidal thoughts” as in the D2S dataset. Moreover, “Suicidal thoughts” samples have also a strong presence which is the result of integrating the D2S dataset in our harvesting process. Since the majority of our samples are coming from self-disclosed users’ tweets, and we apply our DSD model trained on that dataset to the D2S dataset to harvest tweets, our final harvested dataset reflects mainly the distribution of symptoms from the self-disclosed depressed users. However, D2S has some impact, resulting in more samples in the most populated labels of the final harvested dataset.
5.5 Data harvesting in the wild
We use our final model on a bigger set of very loosely related data, but we do not see any increase in accuracy, which suggests that harvesting from irrelevant data is of no use (Sect. 4.2.6).
6 Limitations
-
1.
Our overall dataset size is still small, i.e. for some labels we have a very small amount of data both for training and testing.
-
2.
In the iterative harvesting process we do not employ continuous human annotation or human-in-the-loop strategy since this process requires several such cycles and involving experts in such a framework is also very expensive.
7 Conclusion
We have described a Semi-supervised Learning (SSL) framework, more specifically semi-supervised co-training for gathering depression symptoms data in situ from self-disclosed users’ Twitter timelines. We articulate each step of our data harvesting process and model re-training process. We also discuss our integration of Zero-Shot learning models in this process and their contribution. We show that each of these steps provides moderate to significant accuracy gains. We discuss the effect of harvesting from the samples of an externally curated dataset, and we also try harvesting samples in the wild, i.e., a large noisy dataset with our Final-DSD-Clinician model. In the former case, we find good improvement in the Macro-F1 score. In the latter, we do not see any improvements indicating that there is room for further progress to improve accuracy in those samples. Finally, we discuss the effect of our SSL process for curating small but distributionally relevant samples through both sample distribution and bi-gram distribution for all the labels.
Data availability
The dataset generated and/or analyzed during the current study is not publicly available due to the privacy and ethical implications regarding the identity of Twitter users and tweets. According to Benton et al. (2017) Twitter users may not expect their tweets to have a large audience, that’s why their tweets need to be protected as much as possible. There is also a Twitter policy in place for sharing data, where Twitter discourages sharing tweets directly with third party (Twitter Data Sharing Policy). Therefore, we're still trying to find the best way in which we may share data. We will not release our dataset to any interested party, until they acquire the signed consent form from the external research organizations from which we collected our datasets and created our own, because, we are not allowed to share datasets collected by these organizations.
References
Agrawal, R., & Srikant, R. (1994). Fast algorithms for mining association rules. In Proc. 20th int. conf. very large data bases, VLDB (Vol. 1215, pp. 487–499). Citeseer.
Benton, A., Coppersmith, G., & Dredze, M. (2017). Ethical research protocols for social media health research. In Proceedings of the first ACL workshop on ethics in natural language processing (pp. 94–102).
Boyd, J. H., Weissman, M. M., Thompson, W. D., & Myers, J. K. (1982). Screening for depression in a community sample: Understanding the discrepancies between depression symptom and diagnostic scales. Archives of General Psychiatry, 39(10), 1195–1200.
Coppersmith, G., Dredze, M., Harman, C., Hollingshead, K., & Mitchell, M. (2015). Clpsych 2015 shared task: Depression and ptsd on twitter. In Proceedings of the 2nd workshop on computational linguistics and clinical psychology: From linguistic signal to clinical reality (pp. 31–39).
De Choudhury, M., & De, S. (2014). Mental health discourse on reddit: Self-disclosure, social support, and anonymity. In ICWSM.
De Choudhury, M., Gamon, M., Counts, S., & Horvitz, E. (2013). Predicting depression via social media. In ICWSM (p. 2).
Farruque, N., Goebel, R., Zaïane, O. R., & Sivapalan, S. (2021). Explainable zero-shot modelling of clinical depression symptoms from text. In 2021 20th IEEE international conference on machine learning and applications (ICMLA) (pp. 1472–1477). IEEE.
Farruque, N., Zaiane, O., & Goebel, R. (2019). Augmenting semantic representation of depressive language: From forums to microblogs. In Joint European conference on machine learning and knowledge discovery in databases (pp. 359–375). Springer.
Gowen, K., Deschaine, M., Gruttadara, D., & Markey, D. (2012). Young adults with mental health conditions and social networking websites: Seeking tools to build community. Psychiatric Rehabilitation Journal, 35(3), 245.
Jamil, Z., Inkpen, D., Buddhitha, P., White, K. (2017). Monitoring tweets for depression to detect at-risk users. In Proceedings of the 4th workshop on computational linguistics and clinical psychology—from linguistic signal to clinical reality (pp. 32–40).
Losada, D. E., & Crestani, F. (2016). A test collection for research on depression and language use. In International conference of the cross-language evaluation forum for European languages (pp. 28–39). Springer.
Ma, L., Wang, Z., & Zhang, Y. (2017). Extracting depression symptoms from social networks and web blogs via text mining. In Proceedings of Bioinformatics research and applications: 13th international symposium, ISBRA 2017, Honolulu, HI, USA, 29 May–2 June 2017 (Vol. 13, pp. 325–330). Springer.
McClosky, D., Charniak, E., & Johnson, M. (2006). Effective self-training for parsing. In Proceedings of the human language technology conference of the NAACL, main conference (pp. 152–159).
Mowery, D., Smith, H., Cheney, T., Stoddard, G., Coppersmith, G., Bryan, C., & Conway, M. (2017). Understanding depressive symptoms and psychosocial stressors on Twitter: A corpus-based study. Journal of Medical Internet Research, 19(2), e48.
Mowery, D. L., Park, Y. A., Bryan, C., & Conway, M. (2016). Towards automatically classifying depressive symptoms from Twitter data for population health. In Proceedings of the workshop on computational modeling of people’s opinions, personality, and emotions in social media (PEOPLES) (pp. 182–191).
Naslund, J., Aschbrenner, K., Marsch, L., & Bartels, S. (2016). The future of mental health care: Peer-to-peer support and social media. Epidemiology and Psychiatric Sciences, 25(2), 113–122.
Naslund, J. A., Grande, S. W., Aschbrenner, K. A., & Elwyn, G. (2014). Naturally occurring peer support through social media: The experiences of individuals with severe mental illness using youtube. PLoS ONE, 9(10), 110171.
O’Keeffe, G. S., & Clarke-Pearson, K. (2011). The impact of social media on children, adolescents, and families. Pediatrics, 127(4), 800–804.
Reece, A. G., Reagan, A. J., Lix, K. L., Dodds, P. S., Danforth, C. M., & Langer, E. J. (2017). Forecasting the onset and course of mental illness with twitter data. Scientific Reports, 7(1), 13006.
Rude, S., Gortner, E.-M., & Pennebaker, J. (2004). Language use of depressed and depression-vulnerable college students. Cognition & Emotion, 18(8), 1121–1133.
Safa, R., Bayat, P., & Moghtader, L. (2022). Automatic detection of depression symptoms in Twitter using multimodal analysis. The Journal of Supercomputing, 78(4), 4709–4744.
Seabrook, E. M., Kern, M. L., Fulcher, B. D., & Rickard, N. S. (2018) Predicting depression from language-based emotion dynamics: Longitudinal analysis of Facebook and Twitter status updates. Journal of Medical Internet Research, 20(5), e168.
Shen, G., Jia, J., Nie, L., Feng, F., Zhang, C., Hu, T., Chua, T.-S., Zhu, W. (2017). Depression detection via harvesting social media: A multimodal dictionary learning solution. In IJCAI (pp. 3838–3844).
The classification of depression and depression rating scales/questionnaires. In Depression in adults with a chronic physical health problem: Treatment and management. British Psychological Society (2010)
Tlelo-Coyotecatl, I., Escalante, H. J., & Montes y Gómez, M. (2022) Depression recognition in social media based on symptoms’ detection. Procesamiento del Lenguaje Natural, Revista, 68, 25–37.
Trotzek, M., Koitka, S., & Friedrich, C. M. (2018). Utilizing neural networks and linguistic metadata for early detection of depression indications in text sequences. IEEE Transactions on Knowledge and Data Engineering, 32(3), 588–601.
Vioulès, M. J., Moulahi, B., Azé, J., & Bringay, S. (2018). Detection of suicide-related posts in Twitter data streams. IBM Journal of Research and Development, 62(1), 7–1.
World Health Organization. (2023). Suicide. Retrieved from https://www.who.int/news-room/fact-sheets/detail/suicide
Yadav, S., Chauhan, J., Sain, J.P., Thirunarayan, K., Sheth, A., & Schumm, J. (2020). Identifying depressive symptoms from Tweets: Figurative language enabled multitask learning framework. arXiv preprint. arXiv:2011.06149
Yazdavar, A. H., Al-Olimat, H. S., Ebrahimi, M., Bajaj, G., Banerjee, T., Thirunarayan, K., Pathak, J., & Sheth, A. (2017). Semi-supervised approach to monitoring clinical depressive symptoms in social media. In Proceedings of the 2017 IEEE/ACM international conference on advances in social networks analysis and mining 2017 (pp. 1191–1198). ACM.
Acknowledgements
We are grateful to the annotators for allocating their time for data annotation.
Funding
We are grateful to Alberta Machine Intelligence Institute (AMII), Natural Sciences and Engineering Research Council of Canada (NSERC), and MITACS for their support.
Author information
Authors and Affiliations
Contributions
N.F. developed the original research idea, designed and conducted experiments, annotated samples, and wrote and reviewed the manuscript. R.G. reviewed the manuscript and managed funding. S.S. helped in creating annotation guidelines, annotated samples, and reviewed the manuscript. O.Z. reviewed the manuscript.
Corresponding author
Ethics declarations
Ethical approval
We obtained ethics approval from the University of Alberta’s research ethics office for “Depression Detection from Social Media Language Usage” (Pro00099074), “Depression Dataset Collection” (Pro00082738), and “Social Media Data Annotation by Human” (Pro00091801).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix 1: Apriori rules
Here we provide the strong rules mined from DSD-Clinician-Tweets-Original-Train (Table 22).
Appendix 2: Mental-BERT training configuration for DPD and DSD
Here we report the training configuration for Mental-BERT based DPD and DSD (Table 23).
For DSD we use BCE Loss on the output of last layer of our Mental-BERT model which is based on sigmoid functions for each nodes corresponding to each depression symptoms labels. For DPD, we use BCE loss on the softmaxed output for each binary labels i.e. depression vs control. We do not freeze any layers in our fine-tuning process because it turned out to be detrimental to the model accuracy.
Appendix 3: Annotation guideline
1.1 Social media data annotation by human
For this annotation task, an annotator has to label or classify a social media post (i.e. a tweet) in one or more of the following depression symptom categories which suit best for that social media post through a web tool:
-
1.
Inability to feel pleasure or Anhedonia
-
2.
Low mood
-
3.
Change in sleep pattern
-
4.
Fatigue or loss of energy
-
5.
Weight change or change in appetite
-
6.
Feelings of worthlessness or excessive inappropriate guilt
-
7.
Diminished ability to think or concentrate or indecisiveness
-
8.
Psychomotor agitation or inner tension
-
9.
Psychomotor retardation
-
10.
Suicidal thoughts or self-harm
-
11.
Evidence of clinical depression
-
12.
No evidence of clinical depression
-
13.
Gibberish
Detailed description of these categories with examples are as follows:
The following sections need to be very carefully read to better understand what each category means. We divide the description under each category into three parts: “Lead”, “Elaboration”, and “Example”. “Lead” contains the summary or gist of the symptomatology. “Elaboration” provide a broader description of the symptomatology accompanied by a few relevant “Examples”. These sections have been developed with careful considerations of criteria defined in the DSM-5 and MADRS, BDI, CES-D and PHQ-9 depression rating scales.
1.2 Depression symptoms labels
-
1
Inability to feel pleasure or anhedonia
-
(a)
Lead: Subjective experience of reduced interest in the surroundings or activities, that normally give pleasure.
-
(b)
Elaboration: Dissatisfied and bored about everything. Not enjoying things as one would used to. Not enjoying life. Lost Interest in other people. Lost interest in sex. Can’t cry anymore even though one wants to.
-
(c)
Example:
-
(i)
I feel numb.
-
(ii)
I am dead inside.
-
(iii)
I don’t give a damn to anything anymore.
-
(i)
-
(a)
-
2
Diminished ability to think or concentrate or indecisiveness
-
(a)
Lead: Difficulties in collecting one’s thoughts mounting to incapacitating lack of concentration.
-
(b)
Elaboration: Can’t make decisions at all anymore. Trouble keeping one’s mind on what one was doing. Trouble concentrating on things.
-
(c)
Example:
-
(i)
I can’t make up my mind these days.
-
(i)
-
(a)
-
3
Change in sleep pattern
-
(a)
Lead: Reduced duration or depth of sleep, or increased duration of sleep compared to one’s normal pattern when well.
-
(b)
Elaboration: Trouble falling or staying asleep. Waking up earlier and cannot go back to sleep. Sleep was restless (wake up not feeling rested). Sleeping too much.
-
(c)
Example:
-
(i)
It’s 3 am, and I am still awake.
-
(ii)
I sleep all day!
-
(i)
-
(a)
-
4
Fatigue or loss of energy
-
(a)
Lead: Any physical manifestation of tiredness.
-
(b)
Elaboration: Feeling tired. Insufficient energy for tasks. Feeling too tired to do anything.
-
(c)
Example:
-
(i)
I feel tired all day.
-
(ii)
I feel sleepy all day.
-
(iii)
I get exhausted very easily.
-
(i)
-
(a)
-
5
Feelings of worthlessness or excessive inappropriate guilt
-
(a)
Lead:Representing thoughts of guilt, inferiority, self-reproach, sinfulness, and self-depreciation.
-
(b)
Elaboration: Feeling like a complete failure, Feeling guilty, Feeling of being punished. Self-hate. Disgusted and Disappointed in oneself. Self-blaming for everything bad happens. Believe that one looks ugly or unattractive. Having crying spells. Feeling lonely. People seem unfriendly. Felt like all other people dislike oneself.
-
(c)
Example:
-
(i)
Leave me alone, I want to go somewhere where there is no one.
-
(ii)
I am so alone...
-
(iii)
Everything bad happens, happens because of me.
-
(i)
-
(a)
-
6
Low mood
-
(a)
Lead: Despondency, Gloom, Despair, Depressed Mood, Low Spirits, Feeling of being beyond help without hope.
-
(b)
Elaboration: Feeling down. Feeling sad. Discouraged about future. Hopelessness. Feeling like it’s not possible to shake of the blues even with the help of family and friends.
-
(c)
Example:
-
(i)
Life will never get any better.
-
(ii)
I don’t know why but I feel so empty.
-
(iii)
I am so lost.
-
(iv)
There is no hope to get out of this bad situation.
-
(i)
-
(a)
-
7
Psychomotor agitation or inner tension
-
(a)
Lead: Ill defined discomfort, edginess, inner-turmoil, mental tension mounting to either panic, dread or anguish.
-
(b)
Elaboration: Feeling irritated and annoyed all the time. Bothered by things that usually don’t bother. Feeling fearful. Feeling Restless. Feeling Mental Pain.
-
(c)
Example:
-
(i)
It’s my life so I decide what to do next, mind your own business, don’t bother!
-
(ii)
You have no idea how much pain you gave me!
-
(i)
-
(a)
-
8
Psychomotor retardation or lassitude
-
(a)
Lead: Difficulty getting started or slowness initiating and performing everyday activities.
-
(b)
Elaboration: Feeling everything one do requires effort. Could not get going. Talked less than usual. Have to push oneself to do anything. Everything is a struggle. Moving or talking slowly.
-
(c)
Example:
-
(i)
I don’t feel like moving from the bed.
-
(i)
-
(a)
-
9
Suicidal thoughts or self-Harm
-
(a)
Lead: Feeling of Life is not worth living, suicidal thoughts, preparation for suicide.
-
(b)
Elaboration: Recurrent thoughts of death (not just fear of dying), recurrent suicidal ideation without specific plan, or suicide attempt, or a specific plan for suicide. Thoughts of self-harm. Suicidal ideation. Drug abuse.
-
(c)
Example:
-
(i)
I want to leave for the good.
-
(ii)
0 days clean.
-
(i)
-
(a)
-
10
Weight change or change in appetite
-
(a)
Lead: Loss or gain of appetite or weight than usual.
-
(b)
Elaboration: Increase in weight. Decrease in weight. Increase in appetite. Decrease in appetite. Do not feel like eating. Poor appetite. Loss of desire to food, forcing oneself to eat. Eating a lot but not feeling satiated. Eating even if one is full. Eating in large amount of food quickly and repeatedly. Difficulty in stop eating.
-
(c)
Example:
-
(i)
I think I am over eating these days!
-
(ii)
I don’t feel like eating anything!
-
(i)
-
(a)
-
11
Evidence of clinical depression
-
(a)
Elaboration: Any social media post which do not necessarily fit into any of the above symptoms, however still carry signs of depression or representing many symptoms at a time, so it’s very hard to fit it in a few symptoms.
-
(b)
Example:
-
(i)
I feel like I am drowning …
-
(i)
-
(a)
-
12
No evidence of clinical depression
-
(a)
Elaboration: Political stance or personal opinion, inspirational statement or advice, unsubstantiated claim or fact.
-
(b)
Example:
-
(i)
People who eat dark chocolate are less likely to be depressed.
-
(i)
-
(a)
-
13
Gibberish
-
(a)
Elaboration: If you are not sure what a social media post means i.e. if a social media post does not make sense or it’s gibberish, then annotate it as Gibberish.
-
(a)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Farruque, N., Goebel, R., Sivapalan, S. et al. Depression symptoms modelling from social media text: an LLM driven semi-supervised learning approach. Lang Resources & Evaluation (2024). https://doi.org/10.1007/s10579-024-09720-4
Accepted:
Published:
DOI: https://doi.org/10.1007/s10579-024-09720-4