Abstract
Centromeres are complex structures involved in an evolutionarily conserved function, the correct segregation of chromosomes and chromatids during meiosis and mitosis. The centromere is determined by epigenetic processes that result in a particular nucleosome organization (CEN chromatin) that differs from the rest of the chromatin including the heterochromatin that normally surrounds the centromere in higher organisms. Many of the current models of centromere origin and organization rely on the molecular and cytological characterization of minichromosomes and their derivatives, and on studies on the origin and maintenance of neocentromeres. Here, we describe the peculiar centromere organization observed in In(2Rh)PL, a paracentric D. melanogaster inversion in which the centromere is maintained in its natural context but is directly flanked by a euchromatic domain as a result of the rearrangement. We have identified the breakpoints of the inversion and show that the proximal one is within the centromere region. The data presented suggest that, notwithstanding the loss of all the pericentric 2Rh heterochromatin, the centromere of the In(2Rh)PL chromosome is still active but presents a nucleosomal organization quite different from the organization usually observed in the centromeric region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In many organisms the centromere is embedded within heterochromatin, a compact chromosomal structure that differs structurally from euchromatin, in which most genes reside. Why such an organization of the centromeric region is so common is not completely understood, although several data suggest a role of pericentric chromatin in maintaining the cohesion of chromatids during mitosis and meiosis (Bernard and Allshire 2002; Pidoux and Allshire 2004).
Centromere identity implies the ability to form the kinetochore, the apparatus responsible for binding chromatids to spindle microtubules; it does not appear related to a particular DNA sequence, but rather to a specific modification of the local nucleosomal structure. The protein CENP-A, a variant form of the H3 histone associated with the kinetochore and also present in human neocentromeres (Wong and Choo 2001), represents a specific marker of the centromeric nucleosomal structure. How CENP-A is specifically deposited at the centromere is not completely known. However, recent results on the fission yeast S. pombe show that the pericentromeric regions may be necessary for the deposition of CNP1 (the S. pombe protein orthologous to CENP-A) at the central kinetochore core sequence by the RNA interference machinery that triggers heterochromatin formation (Folco et al. 2008).
Many local histone modifications may occur in chromatin. Methylation, acetylation and phosphorylation of histones are thought to comprise a ‘histone code’ by which particular combinations of histone modifications are linked to an active, permissive or inactive state of the chromatin (Jenuwein and Allis 2001). At the centromere, histone modifications are similar in D. melanogaster and human chromosomes (Blower et al. 2002) and result in separate centromeric and pericentromeric domains where CENP-A-containing nucleosomes are interspersed with nucleosomes containing H3K4me2, and flanked on both sides by nucleosomes containing H3K9me2 (Sullivan and Karpen 2004). H3K9me2 is usually associated with heterochromatic domains able to silence a gene when located within or near it (Lacher et al. 2001; Nakayama et al. 2001), whereas H3K4me2 is also found in transcriptionally permissive chromatin (Jenuwein and Allis 2001). In addition, there is evidence that CEN chromatin may present a specialized nucleosomal structure (Dalal et al. 2007).
Chromosomal rearrangements are widely used in chromatin studies because the rearranged chromosomal organization often allows the characterization of important aspects of heterochromatin biology. For example, D. melanogaster In(1)wm4 has been a critical tool for the study of the silencing effect of heterochromatin on the expression of the white gene (Position Effect Variegation) resulting from the new euchromatin–heterochromatin juxtaposition and, in addition, for the identification of the genes involved in this process (Grewal and Elgin 2002).
Here we present the molecular and cytological characterization of the D. melanogaster In(2Rh)PL paracentric inversion, which determines a variegated eye phenotype due to the insertion of a P(w+) transgene within the centromeric region of chromosome 2. We have mapped the breakpoints of the rearrangement and show that the proximal breakpoint affects the centromere, which in the inverted chromosome displays a novel CEN chromatin configuration.
Materials and methods
Molecular techniques
To identify the insertion site of the P(w+) transgene we performed inverse PCR on genomic In(2Rh)PL DNA digested with Sau3A, using primers designed on sequences at the 5′ and 3′ ends of the P transposon, as described at http://www.fruitfly.org/about/methods/inverse.pcr.html.
Probes of 1 kb from the 53C region were generated by PCR using primers designed on sequences present at FlyBase (http://flybase.net). The primers used to map the breakpoint by PCR (see Fig. 4) were: D5-5U, 5′ TTTTCTCCGTGTGTTTGCAC; D5-5L, 5′ GCAGCATCGAAAAGAAGGAG; D5-6U, 5′ CCGCTTCTGCTTTTGCTATT; D5-6L, 5′ GGGTCCTTTAATTCGCCACT. The probes D1-2, D7-8 and D5.55, used for FISH and extended fibre analysis, were generated by Long Template PCR (Roche) using the following primers: D1U, 5′ AGCGACTGCCAAACGAATGA and D2L, 5′ TTTCCGCTTTCCCTCTTCAC (D1-2, 12.5 kb); D7U, 5′ GATGGAACTTGGAGGTTGGA and D8L, 5′ CTCTTCGGTTGCAGGACTTC (D7-8, 8.2 kb); D5U, 5′ AAGATGCCAACGGACGATAC and D5-6L, described above (D5.55, 15.1 kb).
In-situ hybridization
Mitotic and polytene chromosome preparations and FISH procedures were according to Dimitri (2004). Probes were labelled with Cy5-dUTP or Cy3-dUTP by a nick-translation kit (GE-Healthcare). The Responder probe (Rsp) is a 240 bp XbaI fragment described in Wu et al. (1988). Chromosomes were counterstained with DAPI. Digital images were obtained using an Olympus epifluorescence microscope equipped with a cooled CCD camera. Greyscale images, obtained by separately recording Cy and DAPI fluorescence with specific filters, were pseudo coloured and merged for the final image using Adobe Photoshop software.
Extended chromatin fibre analysis
Chromatin fibres were prepared from neuroblasts of In(2Rh)PL homozygotes exactly as described in Sullivan and Karpen (2004). Slides were processed for immunofluorescence according to Blower et al. 2002. For FISH analysis after indirect immunofluorescence with CID antibodies, slides were re-fixed in 4% formaldehyde for 15 minutes to crosslink antibodies to proteins and then prepared for FISH as described by Sullivan and Warburton (1999). Labelled DNA was used for each slide at 150–250 ng. Hybridization was for 24 h at 37°C. The extension of the overlap of CID- and H3K9me2-containing nucleosomes was measured using the program softWoRx, essentially as described in Sullivan and Karpen (2004).
Results
Origin of In(2Rh)PL41A-B;53C5-9
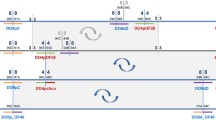
The In(2Rh)PL41A-B;53C5-9 paracentric inversion (hereafter abbreviated In(2Rh)PL) was first described by Berghella and Dimitri (1996) as an x-radiation-induced chromosome 2 rearrangement carrying a P(w+) transgene inserted in the 53C euchromatic region. The inversion places the P(w+) transgene close to the centromeric region and leads to a white-variegated eye phenotype in homozygous flies. In Fig. 1 schematic representations of the original chromosome carrying the P(w+) transgene at 53C and of the In(2Rh)PL chromosome are compared, and the positions of the genetic markers relevant for the present analysis are shown. The centromere region is located in the mitotic heterochromatic h38 band, between the very bright h37 Hoechst band on the left arm (2L) and the much less bright h39 Hoechst band on the right arm (2R) (Gatti and Pimpinelli 1992). Two types of repeated sequences have previously been mapped within the h39 band: the proximal Responder locus (Rsp) (Pimpinelli and Dimitri 1989; Wu et al. 1988) spanning about 600 kb, and, distally, the Bari1 cluster (Caizzi et al. 1993), about 140 kb in size. The centromeric h38 band contains repeats of the AAAGA satellite, which is also found in other heterochromatic bands (Lohe et al. 1993).
Schematic representation of the In(2Rh)PL chromosome. Thin lines represent euchromatin and rectangles represent heterochromatin. (a) Uninverted chromosome with a P(w+) transgene inserted at the 53C region (orange triangle). (b) the x-radiation-induced In(2Rh)PL rearrangement moves the P(w+) transgene near to the centromere (spotted triangle) and is associated with a variegated eye phenotype. C indicates the centromere (mitotic band h38). The flanking heterochromatic h37 and h39 mitotic bands are also indicated, as well as the localization of the main clusters of the Rsp and Bari1 elements in h39
Mapping the proximal breakpoint of In(2Rh)PL
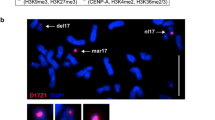
It has been reported that the proximal breakpoint of In(2Rh)PL is located on the left of the Bari1 cluster (Berghella and Dimitri 1996). We extended this analysis by testing whether the Rsp locus is also displaced by the inversion. FISH analysis of mitotic chromosomes clearly demonstrated that the Rsp repeats are displaced by the rearrangement, because the signal observed after hybridization with the Rsp probe is distally located in the inverted chromosome (Fig. 2a). The intensity of the signal is similar in inverted and non-inverted chromosomes, suggesting that, within the resolution limits of the FISH analysis, most or all Rsp repeats have been moved by the rearrangement. Thus, the proximal breakpoint of the inversion appears to fall within the centromeric h38 band.
Localization of Rsp repeats in In(2Rh)PL by FISH analysis. (a) Mitotic chromosomes from a heterozygous In(2Rh)PL/+ male were hybridized with a Rsp probe. The centromere positions (at h38) are indicated by the white lines. Note that in the inverted chromosome (at the bottom) the Rsp signal is located far from the centromere. (b) In polytene chromosomes of In(2Rh)PL homozygotes the 2Rh heterochromatin block is located in the middle of a euchromatic arm (white arrow). The red arrow shows the localization of the P(w+) transgene within the chromocentre
In salivary glands preparations, the heterochromatic regions of all chromosomes usually form a compact, under-replicated and unresolved structure, the chromocentre. However, the 2R heterochromatin of the In(2Rh)PL chromosome is relocated apart from the chromocentre and is observed in the middle of the euchromatic 2R arm (Fig. 2b). As expected, the white probe localizes within the chromocentre.
Mapping the distal breakpoint of In(2Rh)PL
We have mapped the distal breakpoint of In(2Rh)PL as finely as possible to determine the sequences flanking the w+ transgene at 53C also involved in the rearrangement. First, in inverse PCR experiments we localized the P(w+) insertion at position 12 273 876 of the D. melanogaster genomic sequence reported in FlyBase. Starting from the insertion point, we then assayed, by FISH hybridization over mitotic chromosomes of In(2Rh)PL flies, a series of ∼10 kb probes chosen at 10 kb intervals and spanning the 53C region. Figure 3 shows the results obtained with the D1-2 and D7-8 probes. While probe D1-2 gives a signal near the centromere, the signal obtained with probe D7-8 is observed in the original euchromatic location of the corresponding sequence. Thus, the breakpoint must be located between these two probes. Finally, we obtained a more accurate mapping of the breakpoint by FISH hybridization over polytene chromosomes using a series of 1 kb probes from the region identified as including the breakpoint by the analysis described above. The criterion used was that a probe giving a signal over the chromocentre corresponds to a sequence displaced by the inversion, while, conversely, a probe originating a signal in the 53C region should identify a sequence not displaced by the inversion. Thus, we were able to restrict the position of the In(2Rh)PL distal breakpoint to a 1252 bp sequence in the 53C region. Furthermore, PCR analysis allowed a precise mapping of the breakpoint within the 338 bp sequence between genomic coordinates 12 326 828–12 327 166 (Fig. 4). The breakpoint falls in the 7.4 kb intergenic region between the genes CG7848 and CG4409, and thus does not involve the structural sequences of these genes.
Mapping the In(2Rh)PL distal breakpoint by FISH. (a) Schematic diagram showing the position, in a non-inverted chromosome, of the D1-2 and D7-8 probes with respect to the 53C region (grey bar) and to the P(w+) insertion. Orientation with respect to centromere and telomere is also indicated. (b) Merge picture of a FISH analysis using D1-2 (red signal) and D7-8 (green signal) probes over In(2Rh)PL mitotic chromosomes. C marks the position of the centromere
PCR analysis of the distal breakpoint of In(2Rh)PL. (a) Position of the PCR primers used. D5-5 and D5-6 indicate the two parts of a 1252 bp fragment from the 53C region that were utilized in this analysis. (b) Results of the PCR. ‘In’ lanes correspond to genomic DNA from In(2Rh)PL homozygotes; ‘C’ lanes correspond to DNA from control flies carrying non-inverted chromosomes. On the right the 500 bp ladder marker DNA (Biolabs) is shown
Extended chromatin fibre analysis of the chromosome 2 centromere
According to the mapping of the distal breakpoint of the rearrangement (see above), about 53 kb of DNA from the euchromatic region flanking the P(w+) transgene at 53C is directly juxtaposed to the centromeric h38 region in the In(2Rh)PL chromosome. Apparently, this novel pericentric organization, in which all the 2Rh heterochromatin has been removed, still allows centromere functionality, since homozygous In(2Rh)PL flies are vital and fertile, and neuroblast mitosis does not show detectable anomalies (in preparation, see Discussion). To verify that in In(2Rh)PL the centromere is actually contiguous to euchromatic 53C sequences, we performed co-hybridization fibre FISH experiments over interphase chromatin spreads using as probes an anti-CID antibody (CID is the D. melanogaster protein orthologous to human CENP-A) and DNA fragments from the 53C region including the distal breakpoint of the rearrangement. The probes were: (1) D5.55 (FlyBase position 12 312 042–12 327 166), identifying a sequence closely adjacent to the breakpoint; (2) D1-2 (FlyBase position 12 273 909–12 286 486), identifying a sequence located about 50 kb from the breakpoint; (3) BACR06I15. The distal In(2Rh)PL breakpoint splits the 185 kb insert of BACR06I15 into a 53 kb part encompassing the D1-2 and D5.55 probes and a 132 kb region not involved in the rearrangement. Analyzing the three probes individually over 100 fibre preparations, we found an high percentage of DNA signal within CID-containing nucleosomes (Figs. 5 and 6a–c). Furthermore, when BACR06I15 and the D5.55 probes were used in co-hybridization experiments, signals embedded within the CID nucleosomes were detected (Fig. 6d,e). Taken together, these results suggest that the rearrangement does affect the centromeric architecture.
Extended chromatin fibre analysis. Scale bars represent 3 μm. (a–c) Chromatin fibres from neuroblasts of In(2Rh)PL homozygotes were stained with an anti-CID probe (green signal) and with a 12.5 kb DNA probe that identifies the D1-2 sequence from the 53C region (red signal). The D1-2 sequence is included in the In(2Rh)PL inversion. Three independent merge pictures are shown. Yellow arrows mark the position of the DNA probe. (d–g) Colocalization of the D5-55 probe (e, red signal) and BACR06I15 (f, blue signal) between CID nucleosomes (d, green). (g) Is the merge of (d–f)
As previously mentioned, CID/CENP-A nucleosomes are flanked by H3K9me2 nucleosomes in D. melanogaster and human centromeres (Sullivan and Karpen 2004). The deposition of CID/CENP-A at centromere regions is finely regulated, since overexpression of the protein results in mislocalization of the protein to many non-centromeric chromosomal regions in D. melanogaster (Heun et al. 2006) and expansion of CEN chromatin in human chromosomes (Lam et al. 2006). Moreover, in fission yeast, Cnp1 level is sensitive to the amount of H3 and H4 histones (Castillo et al. 2007). In this context we looked at the CEN chromatin status in the In(2Rh)PL chromosome and we found that H3K9me2 nucleosomes may overlap the CID domain. Although quantification of the overlap in 50 fibre preparations revealed a variable extent of the presence of H3K9me2 in the CID domain (Table 1, Fig. 7), our results show that the removal of the 2Rh heterochromatin may create a novel CEN chromatin configuration clearly different from the organization observed in the centromeric region of the non-rearranged chromosome.
Patterns of interspersion of CID and H3K9me2 in In(2Rh)PL fibre FISH preparations. Two different overlaps are shown: in the panels on the left, the H3K9me2 signal overlaps about 50% the CID signal; whereas in the panels on the right, the H3K9me2 signal completely overlaps the CID domain and also spreads in both flanking regions. For comparison, the panel at the bottom shows the pattern of a non-inverted chromosome from L2 cells (see also Sullivan and Karpen 2004)
Discussion
We have investigated the consequences for the structural organization of the centromeric region of D. melanogaster chromosome 2 when, as a consequence of the In(2Rh)PL paracentric inversion, the pericentromeric heterochromatin that flanks the centromere on its right side is removed and the centromeric region is exposed to a euchromatic environment. First, the inversion has been characterized by mapping its breakpoints as accurately as possible. While it was possible to localize the distal breakpoint of the inversion in a short nucleotide sequence because it falls into a sequenced euchromatic region, the proximal breakpoint falls in a region as yet unresolved by sequencing, so only a mapping strategy based on cytogenetic techniques was practicable for its localization. FISH experiments clearly indicated that this breakpoint falls within the h38 mitotic band, where the centromere has been positioned (Fig. 2). Furthermore, extended chromatin analysis clearly demonstrated that sequences recognized by probes from the euchromatic 53C region are localized between CID containing nucleosomes (Figs. 5 and 6), suggesting that the rearrangement affects the chromatin architecture of the centromere region.
The new configuration arising at the centromere as a result of the In(2Rh)PL rearrangement is very unusual when compared with the architecture of a normal centromere, as the natural heterochromatic domain is maintained on the left side of the centromere but the centromeric region is directly juxtaposed to a large euchromatic region on the right side. Unlike the very simple centromere of budding yeast Saccharomyces cerevisiae, the natural centromeres of most eukaryotic chromosomes are flanked on both sides by heterochromatic domains that harbour specific proteins. Several lines of evidence indicate that this conserved structural organization defines a number of functional and structural domains (Blower and Karpen 2001) that apparently do not depend on a specific underlying DNA sequences. Instead, epigenetic mechanisms are thought to be involved in maintaining the nucleosomal organization in both centric and pericentric chromatin. As a rule, CID/CENPA nucleosomes are interspersed with H3K4me2 nucleosomes at the centromere, whereas H3K9me2 nucleosomes are typically found in pericentric chromatin (Sullivan and Karpen 2004).
It has been suggested that pericentric heterochromatin plays an important role in centromere overall organization, either because it is necessary for de novo centromere formation (Folco et al. 2008) or because it provides a boundary separating CEN chromatin from the rest of heterochromatin (Maggert and Karpen 2001). However, several observations imply a dynamic relationship between centromeric and pericentromeric chromatin domains. For example, spreading of CEN chromatin organization over non-centromeric DNA occurs in human artificial chromosomes (Lam et al. 2006); in D. melanogaster a neocentromere can be generated starting from an adjacent active centromere (Maggert and Karpen 2001); CID can be replaced by H3 when depleted by RNAi (Blower et al. 2002); and overexpression of CENPA can lead to its spreading along chromatin fibres (Lam et al. 2006). Moreover, it has been shown that chromosome rearrangements generating a new euchromatin–heterochromatin junction may per se alter the distribution of H3K9me2, inducing new enrichment sites as far away from the breakpoint as several megabases (Yasuhara and Wakimoto 2008).
In this context, our results show that removing the 2Rh heterochromatin from the centromeric region may affect the composition of the CEN chromatin, leading to a ‘heterochromatic’ configuration of the centromeric region as revealed by the presence of H3K9me2 scattered between CID nucleosomes (Fig. 7). At least two events must be envisaged to explain how this novel centromeric configuration may have arisen. First, a silent heterochromatic status may have been induced over the juxtaposed euchromatic 53C region. This could have been triggered either by the reorganized centromeric region itself or by the heterochromatin located on the left side of the centromere. The induced silent chromatin state would have to spread over at least 53 kb of DNA, since the P(w+) transgene located at this distance from the breakpoint still is associated with a variegated phenotype. The second event must have involved the spreading of the CID into the adjacent neoformed heterochromatin as a consequence of the removal by the rearrangement of some natural barrier isolating CEN chromatin from the flanking heterochromatin. The presence of domain boundaries at the centromere has been demonstrated in S. pombe (Scott et al. 2006) and suggested in humans and flies (Sullivan et al. 2001; Maggert and Karpen 2001).
At present, only tentative answers can be given to the crucial interesting questions about the consequences of the new CEN configuration in In(2Rh)PL for the functionality of the kinetochore. The fact that we observed the novel CEN configuration in chromatin fibres from neuroblasts of vital and apparently normal flies suggests strongly that at least in such cells centromere functionality is maintained. Moreover, cytological observation of 500 neuroblast mitoses from flies homozygous for the In(2Rh)PL inversion did not provide any evidence of an increase of mitotic defects (anaphase bridges, aneuploidies, chromosome loss or chromosome fragmentation frequency) in comparison with controls not carrying the rearrangement (in preparation).
Of course, at present it cannot be excluded that in other cells or tissues a different configuration of the centromere of the inverted chromosomes could be present, and that such cells were responsible for the survival and lack of apparent abnormalities of flies carrying the In(2Rh)PL inversion. However, we believe that further, detailed functional and molecular studies of the peculiar centromere architecture reported in this paper might provide interesting insights about the structural organization of the eukaryotic chromosome.
In summary, our data are consistent with the emerging picture of the centromere as a dynamic structure that is able to take into account the state of the surrounding chromatin to assume the compositional architecture required for its function (Dawe and Henikoff 2006; Lam et al. 2006; Castillo et al. 2007).
Abbreviations
- In(2Rh)PL:
-
inversion of chromosome 2 involving heterochromatin of right arm
- BAC:
-
bacterial artificial chromosome
- CCD:
-
cooled camera device
- CID:
-
centromere identifier
- Cy-dUTP:
-
5-amino-propargyl-2′-deoxyuridine 5′-triphosphate-coupled either to Cy3 or Cy5 fluorescent dye
- DAPI:
-
4′,6-diamidino-2-phenylindole dihydrochloride
- FISH:
-
fluorescent in-situ hybridization
- H3K4me2:
-
H3 histone dimethylated at lysine in position 4
- H3K9me2:
-
H3 histone dimethylated at lysine in position 9
- Rsp:
-
responder locus
References
Berghella L, Dimitri P (1996) The heterochromatic rolled gene of Drosophila melanogaster is extensively polytenized and transcriptionally active in the salivary gland chromocentre. Genetics 144:117–125
Bernard P, Allshire RC (2002) Centromeres become unstuck without heterochromatin. Trends Cell Biol 12:419–424
Blower MD, Karpen GH (2001) The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol 3:730–739
Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2:319–330
Caizzi R, Caggese C, Pimpinelli S (1993) Bari-1, a new transposon-like family in Drosophila melanogaster with a unique heterochromatic organization. Genetics 133:335–345
Castillo AC, Mellone BG, Partridge JF et al (2007) Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet 3:e121
Dalal Y, Wang H, Lindsay S, Henikoff S (2007) Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol 5:e218
Dawe RK, Henikoff S (2006) Centromere put epigenetics in the driver’s seat. Trends Biochem Sci 31:662–669
Dimitri P (2004) Fluorescent in situ hybridization with transposable element probes to mitotic chromosomal heterochromatin of Drosophila. Methods Mol Biol 260:29–39
Folco HD, Pidoux AL, Urano T, Allshire RC (2008) Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319:94–97
Gatti M, Pimpinelli S (1992) Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet 26:239–275
Grewal SIS, Elgin SCR (2002) Heterochromatin: new possibilities for inheritance of structure. Curr Opin Genet Dev 12:178–187
Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH (2006) Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10:303–315
Jenuwein T, Allis CD (2001) Translating the histone code. Science 293:1074–1080
Lacher M, O’Carroll D, Rea S Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120
Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA (2006) Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci USA 103:4186–4191
Lohe AR, Hilliker AJ, Roberts PA (1993) Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134:1149–1174
Maggert KA, Karpen GH (2001) The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158:1615–1628
Nakayama J, Rice JC, Stahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113
Pidoux AL, Allshire R (2004) Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res 12:521–534
Pimpinelli S, Dimitri P (1989) Cytogenetic organization of the Rsp (Responder) locus in Drosophila melanogaster. Genetics 121:765–772
Scott KC, Merrett SL, Willard HF (2006) A heterochromatin barrier partitions in fission yeast centromere into discrete domain. Curr Biol 16:119–129
Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Stuct Mol Biol 11:1076–1083
Sullivan B, Warburton P (1999) Studying the progression of vertebrate chromosomes through mitosis by immunofluorescence and FISH. In: Bickmore W (ed) Chromosome structural analysis: a practical approach. IRL Press, Oxford, pp 81–101
Sullivan BA, Blower MD, Karpen GH (2001) Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet 2:584–596
Wong LH, Choo KH (2001) Centromere on the move. Genome Res 11:513–516
Wu CI, Lyttle TW, Wu ML, Lin GF (1988) Association between a satellite DNA sequence and the Responder of Segregation distorter in Drosophila melanogaster. Cell 54:179–189
Yasuhara JC, Wakimoto BT (2008) Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin–heterochromatin transition zones. PLoS Genet 4:e16
Acknowledgements
Sarantis Chlamydas was supported by Italian PhD fellowship. We thank P. D’Addabbo for the preparation of the figures. Financial support was provided by the Ministero dell’Università e della Ricerca (MUR) to RC.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Herbert McGregor.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Chlamydas, S., Heun, P., Dimitri, P. et al. The paracentric inversion In(2Rh)PL alters the centromeric organization of chromosome 2 in Drosophila melanogaster . Chromosome Res 17, 1–9 (2009). https://doi.org/10.1007/s10577-008-9000-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-008-9000-3