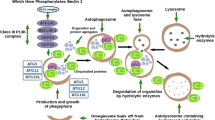
Abstract
Organelles juxtaposition has been detected for decades, although only recently gained importance due to a pivotal role in the regulation of cellular processes dependent on membrane contact sites. Endoplasmic reticulum (ER) and mitochondria interaction is a prime example of organelles contact sites. Mitochondria-associated membranes (MAM) are proposed to harbor ER-mitochondria tether complexes, mainly when these organelles are less than 30 nm apart. Dysfunctions of proteins located at the MAM are associated with neurodegenerative diseases such as Parkinson’s, Alzheimer’s and amyotrophic lateral sclerosis, as well as neurodevelopmental disorders; hence any malfunction in MAM can potentially trigger cell death. This review will focus on the role of ER-mitochondria contact sites, regarding calcium homeostasis, lipid metabolism, autophagy, morphology and dynamics of mitochondria, mainly in the context of neurodegenerative diseases. Approaches that have been employed so far to study organelles contact sites, as well as methods that were not used in neurosciences yet, but are promising and accurate ways to unveil the functions of MAM during neurodegeneration, is also discussed in the present review.


Similar content being viewed by others
Abbreviations
- α-syn:
-
Alpha-synuclein
- Aβ:
-
Amyloid-beta peptide
- ACAT1:
-
Cholesterol acyltransferase/sterol O-acyltransferase 1 (same as SOAT1)
- AD:
-
Alzheimer’s disease
- ALS:
-
Amyotrophic lateral sclerosis
- APEX:
-
Ascorbate peroxidase
- APOE4:
-
Apolipoprotein E4
- APP:
-
Amyloid precursor protein
- ATG2A:
-
Autophagy-related protein 2 homolog A
- ATG9A:
-
Autophagy-related protein 9 homolog A
- ATG14L:
-
Autophagy-related 14-like
- BAP31:
-
B-cell receptor-associated protein 31
- BECN1:
-
Beclin-1
- BiC:
-
Bimolecular complementation
- BioID:
-
Proximity based biotin identification
- BRET:
-
Bioluminescence resonance energy transfer
- CACNA1A:
-
Calcium Voltage-Gated Channel Subunit Alpha1 A
- CE:
-
Cholesteryl-ester
- CLEM:
-
Correlative light and electron microscopy
- CMT2A:
-
Charcot–Marie–Tooth type 2A
- Cryo-ET:
-
Electron cryo-tomography
- Cryo-FIB:
-
Cryo-focused ion beam
- ddFP:
-
Dimerization-dependent fluorescent protein
- DFCP1:
-
Double FYVE domain-containing protein 1
- DGAT2:
-
Diacylglycerol O-acyltransferase 2
- DRP1:
-
Dynamin-Related Protein 1
- EI24:
-
Etoposide-induced protein 2.4
- EM:
-
Electron microscopy
- ER:
-
Endoplasmic reticulum
- FACL4:
-
Fatty acid CoA ligase 4 (same as ACS4)
- FIB-SEM:
-
Focused ion beam-scanning EM
- FIP200:
-
Focal adhesion kinase (FAK) family interacting protein of 200 kDa
- FIS1:
-
Mitochondrial fission 1 protein
- FRET:
-
Fluorescence resonance energy transfer
- GRP75:
-
Glucose regulated protein 75
- GSK-3β:
-
Glycogen synthase kinase-3 beta
- hiPS:
-
Human-induced pluripotent stem cell
- IP3R:
-
Inositol 1, 4, 5-trisphosphate receptor
- KIF5C:
-
Kinesin Family Member 5C
- LB:
-
Lewy bodies
- LRRK2:
-
Leucine-rich repeat kinase 2
- MAM:
-
Mitochondria-associated membranes
- MEF:
-
Mouse embryonic fibroblasts
- MERCs:
-
Mitochondria-ER contact sites
- MFN1 and MFN2:
-
Mitochondrial fusion GTPases mitofusins 1 and 2
- MID49:
-
Mitochondrial dynamics protein of 49 kDa
- MIRO1 and 2:
-
Mitochondrial Rho GTPase 1 and 2 (same as RHOT1 and 2)
- MITOL:
-
Mitochondrial ubiquitin ligase (same as MARCH5)
- Mul1:
-
Mitochondrial E3 ubiquitin protein ligase 1
- ORP5/8:
-
Oxysterol-binding protein related-protein 5 and 8
- PC:
-
Phosphatidylcholine
- PD:
-
Parkinson’s disease
- PDZD8:
-
PDZ domain-containing protein 8
- PE:
-
Phosphatidylethanolamine
- PEMT2:
-
Phosphatidylethanolamine N-methyltransferase 2
- PINK1:
-
PTEN-induced kinase 1
- PLA:
-
Proximity ligation assay
- PSEN 1 and 2:
-
Presenilin 1 and 2
- PSS 1 and 2:
-
Phosphatidylserine synthases 1 and 2
- PTPIP51:
-
Protein tyrosine phosphatase interacting protein 51 (same as RMDN3)
- ROS:
-
Reactive oxygen species
- SBF-SEM:
-
Serial block-face scanning electron microscopy
- SEM:
-
Scanning electron microscopy
- Sig-1R:
-
Sigma-1 receptor
- SNCA:
-
Alpha-synuclein gene
- TDP-43:
-
TAR DNA binding protein 43
- TEM:
-
Transmission electron microscopy
- TRAK:
-
Trafficking kinesin protein (same as Milton)
- ULK1:
-
Unc-51 like autophagy activating kinase
- VAPB:
-
Vesicle-associated membrane protein (VAMP)-associated protein B
- VDAC:
-
Voltage-dependent anion-selective channel protein 1
- VPS13A:
-
Vacuolar Protein Sorting 13 Homolog A
References
AlSaif A, AlMohanna F, Bohlega S (2011) A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol 70:913–919. https://doi.org/10.1002/ana.22534
Alford SC, Ding Y, Simmen T, Campbell RE (2012) Dimerization-dependent green and yellow fluorescent proteins. ACS Synth Biol 1:569–575. https://doi.org/10.1021/sb300050j
Alza NP, Iglesias Gonzalez PA, Conde MA, Uranga RM, Salvador GA (2019) Lipids at the crossroad of alpha-synuclein function and dysfunction: biological and pathological implications. Front Cell Neurosci 13:175. https://doi.org/10.3389/fncel.2019.00175
Arakhamia T et al (2020) Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell 180:633–644. https://doi.org/10.1016/j.cell.2020.01.027
Area-Gomez E et al (2018) A key role for MAM in mediating mitochondrial dysfunction in alzheimer disease. Cell Death Dis 9:335. https://doi.org/10.1038/s41419-017-0215-0
Area-Gomez E et al (2012) Upregulated function of mitochondria-associated ER membranes in alzheimer disease. EMBO J 31:4106–4123. https://doi.org/10.1038/emboj.2012.202
Basso V et al (2018) Regulation of ER-mitochondria contacts by Parkin via Mfn2. Pharmacol Res 138:43–56. https://doi.org/10.1016/j.phrs.2018.09.006
Begemann I, Galic M (2016) Correlative light electron microscopy: connecting synaptic structure and function. Front Synaptic Neurosci 8:28. https://doi.org/10.3389/fnsyn.2016.00028
Bento CF et al (2016) Mammalian autophagy: how does it work? Annu Rev Biochem 85:685–713. https://doi.org/10.1146/annurev-biochem-060815-014556
Berenguer-Escuder C et al (2020) Impaired mitochondrial-endoplasmic reticulum interaction and mitophagy in miro1-mutant neurons in parkinson’s disease. Hum Mol Genet 29:1353–1364. https://doi.org/10.1093/hmg/ddaa066
Berenguer-Escuder C et al (2019) Variants in miro1 cause alterations of ER-mitochondria contact sites in fibroblasts from parkinson’s disease patients. J Clin Med. https://doi.org/10.3390/jcm8122226
Bernard-Marissal N et al (2019) Altered interplay between endoplasmic reticulum and mitochondria in charcot-marie-tooth type 2A neuropathy. Proc Natl Acad Sci U S A 116:2328–2337. https://doi.org/10.1073/pnas.1810932116
Bernhard W, Haguenau F, Gautier A, Oberling C (1952) Submicroscopical structure of cytoplasmic basophils in the liver, pancreas and salivary gland; study of ultrafine slices by electron microscope. Z Zellforsch Mikrosk Anat 37:281–300
Bernhard W, Rouiller C (1956) Close topographical relationship between mitochondria and ergastoplasm of liver cells in a definite phase of cellular activity. J Biophys Biochem Cytol 2:73–78. https://doi.org/10.1083/jcb.2.4.73
Boassa D et al (2019) Split-miniSOG for spatially detecting intracellular protein-protein interactions by correlated light and electron microscopy. Cell Chem Biol 26:1407–1416. https://doi.org/10.1016/j.chembiol.2019.07.007
Cali T, Ottolini D, Negro A, Brini M (2013) Enhanced parkin levels favor ER-mitochondria crosstalk and guarantee Ca(2+) transfer to sustain cell bioenergetics. Biochim Biophys Acta 1832:495–508. https://doi.org/10.1016/j.bbadis.2013.01.004
Cieri D et al (2018) SPLICS: a split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ 25:1131–1145. https://doi.org/10.1038/s41418-017-0033-z
Cohen S, Valm AM, Lippincott-Schwartz J (2018) Interacting organelles. Curr Opin Cell Biol 53:84–91. https://doi.org/10.1016/j.ceb.2018.06.003
Collado J, Fernandez-Busnadiego R (2017) Deciphering the molecular architecture of membrane contact sites by cryo-electron tomography. Biochim Biophys Acta Mol Cell Res 1864:1507–1512. https://doi.org/10.1016/j.bbamcr.2017.03.009
Csordas G et al (2010) Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell 39:121–132. https://doi.org/10.1016/j.molcel.2010.06.029
de Brito OM, Scorrano L (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456:605–610. https://doi.org/10.1038/nature07534
De Vos KJ et al (2012) VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum Mol Genet 21:1299–1311. https://doi.org/10.1093/hmg/ddr559
DeJesus-Hernandez M et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256. https://doi.org/10.1016/j.neuron.2011.09.011
Dorszewska J, Prendecki M, Oczkowska A, Dezor M, Kozubski W (2016) Molecular basis of familial and sporadic alzheimer’s disease. Curr Alzheimer Res 13:952–963. https://doi.org/10.2174/1567205013666160314150501
Elgass KD, Smith EA, LeGros MA, Larabell CA, Ryan MT (2015) Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J Cell Sci 128:2795–2804. https://doi.org/10.1242/jcs.169136
Fanning S et al (2019) Lipidomic analysis of alpha-synuclein neurotoxicity identifies stearoyl CoA desaturase as a target for parkinson treatment. Mol Cell 73:1001–1014. https://doi.org/10.1016/j.molcel.2018.11.028
Fischer TD, Dash PK, Liu J, Waxham MN (2018) Morphology of mitochondria in spatially restricted axons revealed by cryo-electron tomography. PLoS Biol 16:e2006169. https://doi.org/10.1371/journal.pbio.2006169
Fitzpatrick AWP et al (2017) Cryo-EM structures of tau filaments from alzheimer’s disease. Nature 547:185–190. https://doi.org/10.1038/nature23002
Flis VV, Daum G (2013) Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a013235
Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK (2011) ER tubules mark sites of mitochondrial division. Science 334:358–362. https://doi.org/10.1126/science.1207385
Gelmetti V et al (2017) PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy 13:654–669. https://doi.org/10.1080/15548627.2016.1277309
Goedert M, Spillantini MG (2006) A century of alzheimer’s disease. Science 314:777–781. https://doi.org/10.1126/science.1132814
Golovko MY, Rosenberger TA, Feddersen S, Faergeman NJ, Murphy EJ (2007) Alpha-synuclein gene ablation increases docosahexaenoic acid incorporation and turnover in brain phospholipids. J Neurochem 101:201–211. https://doi.org/10.1111/j.1471-4159.2006.04357.x
Gomez-Suaga P, Bravo-San Pedro JM, Gonzalez-Polo RA, Fuentes JM, Niso-Santano M (2018) ER-mitochondria signaling in parkinson’s disease. Cell Death Dis 9:337. https://doi.org/10.1038/s41419-017-0079-3
Gomez-Suaga P, Paillusson S, Stoica R, Noble W, Hanger DP, Miller CCJ (2017) The ER-mitochondria tethering complex VAPB-PTPIP51 regulates autophagy. Curr Biol 27:371–385. https://doi.org/10.1016/j.cub.2016.12.038
Gomez-Suaga P et al (2019) The VAPB-PTPIP51 endoplasmic reticulum-mitochondria tethering proteins are present in neuronal synapses and regulate synaptic activity. Acta Neuropathol Commun 7:35. https://doi.org/10.1186/s40478-019-0688-4
Grossmann D et al (2019) Mutations in RHOT1 disrupt endoplasmic reticulum-mitochondria contact sites interfering with calcium homeostasis and mitochondrial dynamics in parkinson’s disease. Antioxid Redox Signal 31:1213–1234. https://doi.org/10.1089/ars.2018.7718
Guardia-Laguarta C et al (2014) alpha-Synuclein is localized to mitochondria-associated ER membranes. J Neurosci 34:249–259. https://doi.org/10.1523/JNEUROSCI.2507-13.2014
Guardia-Laguarta C, Area-Gomez E, Schon EA, Przedborski S (2015) A new role for alpha-synuclein in parkinson’s disease: alteration of ER-mitochondrial communication. Mov Disord 30:1026–1033. https://doi.org/10.1002/mds.26239
Guo Q et al (2018) In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell 172:696–705. https://doi.org/10.1016/j.cell.2017.12.030
Hamasaki M et al (2013) Autophagosomes form at ER-mitochondria contact sites. Nature 495:389–393. https://doi.org/10.1038/nature11910
Hayashi T, Su TP (2007) Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131:596–610. https://doi.org/10.1016/j.cell.2007.08.036
Hedskog L et al (2013) Modulation of the endoplasmic reticulum-mitochondria interface in alzheimer’s disease and related models. Proc Natl Acad Sci U S A 110:7916–7921. https://doi.org/10.1073/pnas.1300677110
Hertlein V et al (2020) MERLIN: a novel BRET-based proximity biosensor for studying mitochondria-ER contact sites. Life Sci Alliance. https://doi.org/10.26508/lsa.201900600
Hirabayashi Y et al (2017) ER-mitochondria tethering by PDZD8 regulates Ca(2+) dynamics in mammalian neurons. Science 358:623–630. https://doi.org/10.1126/science.aan6009
Hirabayashi Y, Tapia JC, Polleux F (2018) Correlated light-serial scanning electron microscopy (CoLSSEM) for ultrastructural visualization of single neurons in vivo. Sci Rep 8:14491. https://doi.org/10.1038/s41598-018-32820-5
Honrath B, Metz I, Bendridi N, Rieusset J, Culmsee C, Dolga AM (2017) Glucose-regulated protein 75 determines ER-mitochondrial coupling and sensitivity to oxidative stress in neuronal cells. Cell Death Discov 3:17076. https://doi.org/10.1038/cddiscovery.2017.76
Hu F, Zeng C, Long R, Miao Y, Wei L, Xu Q, Min W (2018) Supermultiplexed optical imaging and barcoding with engineered polyynes. Nat Methods 15:194–200. https://doi.org/10.1038/nmeth.4578
Hung V et al (2017) Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 6:e24463. https://doi.org/10.7554/eLife.24463
Jing J, Liu G, Huang Y, Zhou Y (2019) A molecular toolbox for interrogation of membrane contact sites. J Physiol 9:1725–1739. https://doi.org/10.1113/JP277761
Kakimoto Y, Tashiro S, Kojima R, Morozumi Y, Endo T, Tamura Y (2018) Visualizing multiple inter-organelle contact sites using the organelle-targeted split-GFP system. Sci Rep 8:6175. https://doi.org/10.1038/s41598-018-24466-0
Kalia LV, Lang AE (2015) Parkinson’s disease. Lancet 386:896–912. https://doi.org/10.1016/S0140-6736(14)61393-3
Kimura S, Noda T, Yoshimori T (2008) Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct 33:109–122. https://doi.org/10.1247/csf.08005
Kojima R, Endo T, Tamura Y (2016) A phospholipid transfer function of ER-mitochondria encounter structure revealed in vitro. Sci Rep 6:30777. https://doi.org/10.1038/srep30777
Lee JH, Han JH, Kim H, Park SM, Joe EH, Jou I (2019) Parkinson’s disease-associated LRRK2-G2019S mutant acts through regulation of SERCA activity to control ER stress in astrocytes. Acta Neuropathol Commun 7:68. https://doi.org/10.1186/s40478-019-0716-4
Lee KS, Huh S, Lee S, Wu Z, Kim AK, Kang HY, Lu B (2018) Altered ER-mitochondria contact impacts mitochondria calcium homeostasis and contributes to neurodegeneration in vivo in disease models. Proc Natl Acad Sci U S A 115:E8844–E8853. https://doi.org/10.1073/pnas.1721136115
Lee S, Min KT (2018) The interface between ER and mitochondria: molecular compositions and functions. Mol Cells 41:1000–1007. https://doi.org/10.14348/molcells.2018.0438
Lee S, Wang W, Hwang J, Namgung U, Min KT (2019) Increased ER-mitochondria tethering promotes axon regeneration. Proc Natl Acad Sci U S A 116:16074–16079. https://doi.org/10.1073/pnas.1818830116
Lewin TM, Van Horn CG, Krisans SK, Coleman RA (2002) Rat liver acyl-CoA synthetase 4 is a peripheral-membrane protein located in two distinct subcellular organelles, peroxisomes, and mitochondrial-associated membrane. Arch Biochem Biophys 404:263–270. https://doi.org/10.1016/s0003-9861(02)00247-3
Lewis A, Tsai SY, Su TP (2016) Detection of isolated mitochondria-associated ER membranes using the sigma-1 receptor methods. Mol Biol 1376:133–140. https://doi.org/10.1007/978-1-4939-3170-5_11
Lippens S, Kremer A, Borghgraef P, Guerin CJ (2019) Serial block face-scanning electron microscopy for volume electron microscopy Methods. Cell Biol 152:69–85. https://doi.org/10.1016/bs.mcb.2019.04.002
Liu Y, Ma X, Fujioka H, Liu J, Chen S, Zhu X (2019) DJ-1 regulates the integrity and function of ER-mitochondria association through interaction with IP3R3-Grp75-VDAC1. Proc Natl Acad Sci U S A 116:25322–25328. https://doi.org/10.1073/pnas.1906565116
Ma JH et al (2017) Comparative proteomic analysis of the mitochondria-associated ER membrane (MAM) in a long-term type 2 diabetic rodent model. Sci Rep 7:2062. https://doi.org/10.1038/s41598-017-02213-1
Menzies FM et al (2017) Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93:1015–1034. https://doi.org/10.1016/j.neuron.2017.01.022
Mesa-Herrera F, Taoro-Gonzalez L, Valdes-Baizabal C, Diaz M, Marin R (2019) Lipid and lipid raft alteration in aging and neurodegenerative diseases: a window for the development of new biomarkers. Int J Mol Sci 20:3810. https://doi.org/10.3390/ijms20153810
Miller KE, Kim Y, Huh WK, Park HO (2015) Bimolecular fluorescence complementation (BiFC) analysis: advances and recent applications for genome-wide interaction studies. J Mol Biol 427:2039–2055. https://doi.org/10.1016/j.jmb.2015.03.005
Modi S et al (2019) Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery. Nat Commun 10:4399. https://doi.org/10.1038/s41467-019-12382-4
Moustaqim-Barrette A, Lin YQ, Pradhan S, Neely GG, Bellen HJ, Tsuda H (2014) The amyotrophic lateral sclerosis 8 protein, VAP, is required for ER protein quality control. Hum Mol Genet 23:1975–1989. https://doi.org/10.1093/hmg/ddt594
Nagashima S et al (2019) MITOL deletion in the brain impairs mitochondrial structure and ER tethering leading to oxidative stress. Life Sci Alliance 2:e201900308. https://doi.org/10.26508/lsa.201900308
Ottolini D, Cali T, Negro A, Brini M (2013) The Parkinson disease-related protein DJ-1 counteracts mitochondrial impairment induced by the tumour suppressor protein p53 by enhancing endoplasmic reticulum-mitochondria tethering. Hum Mol Genet 22:2152–2168. https://doi.org/10.1093/hmg/ddt068
Paillusson S et al (2017) Alpha-synuclein binds to the ER-mitochondria tethering protein VAPB to disrupt Ca(2+) homeostasis and mitochondrial ATP production. Acta Neuropathol 134:129–149. https://doi.org/10.1007/s00401-017-1704-z
Paillusson S, Stoica R, Gomez-Suaga P, Lau DHW, Mueller S, Miller T, Miller CCJ (2016) There’s something wrong with my MAM; the ER-mitochondria axis and neurodegenerative diseases. Trends Neurosci 39:146–157. https://doi.org/10.1016/j.tins.2016.01.008
Pera M et al (2017) Increased localization of APP-C99 in mitochondria-associated ER membranes causes mitochondrial dysfunction in alzheimer disease. EMBO J 36:3356–3371. https://doi.org/10.15252/embj.201796797
Pizzo P, Pozzan T (2007) Mitochondria-endoplasmic reticulum choreography: structure and signaling dynamics. Trends Cell Biol 17:511–517. https://doi.org/10.1016/j.tcb.2007.07.011
Puglielli L et al (2001) Acyl-coenzyme A: cholesterol acyltransferase modulates the generation of the amyloid beta-peptide. Nat Cell Biol 3:905–912. https://doi.org/10.1038/ncb1001-905
Puri R, Cheng XT, Lin MY, Huang N, Sheng ZH (2019) Mul1 restrains parkin-mediated mitophagy in mature neurons by maintaining ER-mitochondrial contacts. Nat Commun 10:3645. https://doi.org/10.1038/s41467-019-11636-5
Rizzuto R et al (1998) Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280:1763–1766. https://doi.org/10.1126/science.280.5370.1763
Rodriguez-Arribas M et al (2017) Mitochondria-associated membranes (MAMs): overview and its role in parkinson’s disease. Mol Neurobiol 54:6287–6303. https://doi.org/10.1007/s12035-016-0140-8
Rosen DR et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62. https://doi.org/10.1038/362059a0
Rowland AA, Voeltz GK (2012) Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol 13:607–625. https://doi.org/10.1038/nrm3440
Rubinsztein DC, Shpilka T, Elazar Z (2012) Mechanisms of autophagosome biogenesis. Curr Biol 22:R29-34. https://doi.org/10.1016/j.cub.2011.11.034
Schaffer M et al (2019) A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native caenorhabditis elegans tissue. Nat Methods 16:757–762. https://doi.org/10.1038/s41592-019-0497-5
Scorrano L et al (2019) Coming together to define membrane contact sites. Nat Commun 10:1287. https://doi.org/10.1038/s41467-019-09253-3
Sepulveda-Falla D et al (2014) Familial alzheimer’s disease-associated presenilin-1 alters cerebellar activity and calcium homeostasis. J Clin Invest 124:1552–1567. https://doi.org/10.1172/JCI66407
Shu X et al (2011) A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol 9:e1001041. https://doi.org/10.1371/journal.pbio.1001041
Soderberg O et al (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000. https://doi.org/10.1038/nmeth947
Spence EF, Dube S, Uezu A, Locke M, Soderblom EJ, Soderling SH (2019) In vivo proximity proteomics of nascent synapses reveals a novel regulator of cytoskeleton-mediated synaptic maturation. Nat Commun 10:386. https://doi.org/10.1038/s41467-019-08288-w
Sreedharan J et al (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319:1668–1672. https://doi.org/10.1126/science.1154584
Stacchiotti A, Favero G, Lavazza A, Garcia-Gomez R, Monsalve M, Rezzani R (2018) Perspective: mitochondria-ER contacts in metabolic cellular stress assessed by microscopy. Cells 8:5. https://doi.org/10.3390/cells8010005
Stoica R et al (2014) ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun 5:3996. https://doi.org/10.1038/ncomms4996
Stoica R et al (2016) ALS/FTD-associated FUS activates GSK-3beta to disrupt the VAPB-PTPIP51 interaction and ER-mitochondria associations. EMBO Rep 17:1326–1342. https://doi.org/10.15252/embr.201541726
Stone SJ, Levin MC, Zhou P, Han J, Walther TC, Farese RV Jr (2009) The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J Biol Chem 284:5352–5361. https://doi.org/10.1074/jbc.M805768200
Stone SJ, Vance JE (2000) Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated. J Biol Chem 275:34534–34540. https://doi.org/10.1074/jbc.M002865200
Sugiura A et al (2013) MITOL regulates endoplasmic reticulum-mitochondria contacts via mitofusin2. Mol Cell 51:20–34. https://doi.org/10.1016/j.molcel.2013.04.023
Tambini MD et al (2016) ApoE4 upregulates the activity of mitochondria-associated ER membranes. EMBO Rep 17:27–36. https://doi.org/10.15252/embr.201540614
Tang Z et al (2019) TOM40 targets Atg2 to mitochondria-associated ER membranes for phagophore expansion. Cell Rep 28(1744–1757):e1745. https://doi.org/10.1016/j.celrep.2019.07.036
Tao CL et al (2018) Differentiation and characterization of excitatory and inhibitory synapses by cryo-electron tomography and correlative microscopy. J Neurosci 38:1493–1510. https://doi.org/10.1523/JNEUROSCI.1548-17.2017
Tatsuta T, Langer T (2017) Intramitochondrial phospholipid trafficking. Biochim Biophys Acta Mol Cell Biol Lipids 1862:81–89. https://doi.org/10.1016/j.bbalip.2016.08.006
Tocheva EI, Li Z, Jensen GJ (2010) Electron cryotomography. Cold Spring Harb Perspect Biol 2:a003442. https://doi.org/10.1101/cshperspect.a003442
Toyofuku T, Okamoto Y, Ishikawa T, Sasawatari S, Kumanogoh A (2020) LRRK2 regulates endoplasmic reticulum-mitochondrial tethering through the PERK-mediated ubiquitination pathway. EMBO J 39:e100875. https://doi.org/10.15252/embj.2018100875
Turner MR et al (2013) Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol 12:310–322. https://doi.org/10.1016/S1474-4422(13)70036-X
Valadas JS et al (2018) ER lipid defects in neuropeptidergic neurons impair sleep patterns in parkinson’s disease. Neuron 98(1155–1169):e1156. https://doi.org/10.1016/j.neuron.2018.05.022
van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124. https://doi.org/10.1038/nrm2330
van Vliet AR, Verfaillie T, Agostinis P (2014) New functions of mitochondria associated membranes in cellular signaling. Biochim Biophys Acta 1843:2253–2262. https://doi.org/10.1016/j.bbamcr.2014.03.009
Vance JE (1990) Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem 265:7248–7256
Veyrat-Durebex C et al (2019) Metabo-lipidomics of fibroblasts and mitochondrial-endoplasmic reticulum extracts from ALS patients shows alterations in purine, pyrimidine, energetic, and phospholipid metabolisms. Mol Neurobiol 56:5780–5791. https://doi.org/10.1007/s12035-019-1484-7
Vicario M, Cieri D, Brini M, Cali T (2018) The close encounter between alpha-synuclein and mitochondria. Front Neurosci 12:388. https://doi.org/10.3389/fnins.2018.00388
Watanabe S et al (2016) Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol Med 8:1421–1437. https://doi.org/10.15252/emmm.201606403
Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P (2009) Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc 4:1582–1590. https://doi.org/10.1038/nprot.2009.151
Wu Y, Whiteus C, Xu CS, Hayworth KJ, Weinberg RJ, Hess HF, De Camilli P (2017) Contacts between the endoplasmic reticulum and other membranes in neurons. Proc Natl Acad Sci U S A 114:E4859–E4867. https://doi.org/10.1073/pnas.1701078114
Xu S et al (2016) Mitochondrial E3 ubiquitin ligase MARCH5 controls mitochondrial fission and cell sensitivity to stress-induced apoptosis through regulation of MiD49 protein. Mol Biol Cell 27:349–359. https://doi.org/10.1091/mbc.E15-09-0678
Yeshaw WM et al (2019) Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. Elife 8:e43561. https://doi.org/10.7554/eLife.43561
Yuan L, Liu Q, Wang Z, Hou J, Xu P (2019) EI24 tethers endoplasmic reticulum and mitochondria to regulate autophagy flux. Cell Mol Life Sci 8:1591–1606. https://doi.org/10.1007/s00018-019-03236-9
Zampese E, Fasolato C, Kipanyula MJ, Bortolozzi M, Pozzan T, Pizzo P (2011) Presenilin 2 modulates endoplasmic reticulum (ER)-mitochondria interactions and Ca2+ cross-talk. Proc Natl Acad Sci U S A 108:2777–2782. https://doi.org/10.1073/pnas.1100735108
Zhao YG, Liu N, Miao G, Chen Y, Zhao H, Zhang H (2018) The ER contact proteins VAPA/B interact with multiple autophagy proteins to modulate autophagosome biogenesis. Curr Biol 28(1234–1245):e1234. https://doi.org/10.1016/j.cub.2018.03.002
Zhao YG, Zhao H, Miao L, Wang L, Sun F, Zhang H (2012) The p53-induced gene Ei24 is an essential component of the basal autophagy pathway. J Biol Chem 287:42053–42063. https://doi.org/10.1074/jbc.M112.415968
Acknowledgements
Authors were awarded with research grants from Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (FAPESP #13/08028-1, #18/07592-4 and #19/01290-9).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Raeisossadati, R., Ferrari, M.F.R. Mitochondria-ER Tethering in Neurodegenerative Diseases. Cell Mol Neurobiol 42, 917–930 (2022). https://doi.org/10.1007/s10571-020-01008-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-020-01008-9