Abstract
The structure of the fibre network in tissue paper can be complex and difficult to analyze, due to the presence of superimposed structures such as creping and patterns that occur, for example, in through-air-dried (TAD) tissue. Properties like high absorbency, a pleasant handfeel, and strength-related characteristics are closely related to the fibre network structure. Therefore, in addition to standard tissue testing methods, techniques that provide insights into the intrinsic properties of the tissue fibre network are essential for a deeper understanding and potential for further optimization. In this study, we utilized 2D cross-sectional images and 3D X-ray microtomography (\(\mu\)-CT) to evaluate and quantify the intrinsic properties of highly structured TAD tissue. We compared the results obtained from these two methods, focusing on intrinsic thickness, porosity, and the fibre volume to fibre surface area (Fv/Fs) ratio. The open structure of the fibre network, fabric patterns, creping, and protruding fibres make it challenging to define bulk boundaries. Therefore, we examined the effect of different bulk expansion diameters on intrinsic properties. This procedure allows to quantify the effects of under- and overestimation of bulk boundaries, and to determine which regions within the fibre network are affected by bulk expansion. In terms of intrinsic thickness, both 2D and 3D evaluations show similar trends, which facilitates direct comparison of 2D and 3D data. Porosity, on the other hand, does not show any correlation between 2D and 3D-based data. Together with the Fv/Fs parameter, this leads to the conclusion that the depiction of 2D data does not represent the whole fibrous material but predominantly fibres perpendicular or close to perpendicular to the cut plane, whereas 3D data represents all fibres, fibre bonds and network connectivity. This work aims at presenting modern approaches and novel procedures to quantify intrinsic properties of open fibre structures such as tissue, but could also be applied to fibrous networks in general. The introduced methods could provide the basis for future research on the interrelations between intrinsic properties and key tissue properties such as absorbency and handfeel.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, significant efforts have been made to holistically understand and characterize the network structure of cellulose fibre based materials. Regardless of grades considered, such as writing and packaging papers, nonwoven-based products or biodegradable hygiene papers such as tissue, they all have a complex and difficult accessible structure in common. Compared to other grades, tissue products used on a daily basis like kitchen towels, bathroom paper or napkins and handkerchiefs differ greatly in terms of fibre network structure. A low basis weight combined with a high required bulk results in a very open network structure, not comparable to writing or packaging paper, where strength and optical properties are of predominant interest. Softness, water absorbency and wet strength are key properties for tissue which, among other things, strongly depend on fibre network properties such as porosity (Kullander et al. 2012; de Assis et al. 2018; Pawlak et al. 2022). Thus a better understanding of the intrinsic properties of tissue paper networks would be beneficial.
Final tissue characteristics depend mainly on the fibre furnish and machine technology used (de Assis et al. 2018). Typically, a blend of virgin softwood (SW) and hardwood (HW) pulp fibres is used for high quality tissue, but recycled and non-wood fibres are playing an increasingly important role as an alternative fibre resource (de Assis et al. 2019; Zambrano et al. 2021; Kumar et al. 2022). Modern tissue machine concepts can be divided in dry-creped tissue (DCT), through-air-dried (TAD) and hybrid technologies. The TAD technology minimizes mechanical dewatering and uses solely hot air to dry the tissue web prior to the Yankee cylinder and creping. TAD tissue requires a high amount of thermal energy, which is why this technology is mainly used for high quality grades with high bulk requirements (de Assis et al. 2018). TAD tissues have textured surfaces, caused by the wet moulding effect during production. The pattern and topography of the TAD fabric has a major influence on the resulting tissue properties in terms of bulk, stiffness and water absorption and is optimized in various ways (McCabe 2011). Due to the increased complexity of TAD tissue with several superimposed structures such as formation, crepe and patterns, the proper characterization of the tissue bulk properties is challenging.
Standard measurements of tissue bulk properties are caliper (DIN EN ISO 12625-3), basis weight (DIN EN ISO 12625-6) and the resulting apparent bulk/density. From these properties the bulk porosity is often estimated using the density of cellulose fibres (Vieira et al. 2022; Stankovská et al. 2020; Morais et al. 2022). Especially the use of the caliper as basis for subsequent parameters can be considered critical, since the caliper does not represent the actual thickness of structured tissue such as, for instance, TAD (Sung et al. 2005). Machado Charry et al. (2018) introduced the concept of intrinsic properties to differentiate between properties based on measurements inside and outside of the bulk. The measured caliper of the structured tissue under load for instance, is far higher than the mean intrinsic thickness of the fibre network. To access the actual bulk structure, measurements which depict the cross section of tissue paper are inevitable. In the following several destructive and non-destructive 2D and 3D methods recently applied in paper structure analysis are introduced.
In the field of low basis weight papers, such as tissue, a study regarding the non-destructive 2D cross section evaluation was recently performed by Agarwal et al. (2023). They developed an optical method for high resolution cross-sectional edge images with a subsequent evaluation of the centerline to determine the crepe index of dry-creped tissue. However, destructive methods for cross-sectional evaluation are more common. In particular, scanning electron microscopy (SEM) has been widely used despite the quite laborious material preparation such as coating, cutting and critical measuring environment (cooling). Das et al. (2020) combined optical cross section images with SEM data to determine the dominant crepe wavelength in DCT. Cross-section SEM images were qualitatively used by de Assis et al. (2020) to show the effect of compressive forces at the creping blade on fibre web structure. A quantitative evaluation based on SEM was done by Vieira et al. (2022) to determine the crepe wave height impacted by mechanical compression by embossing. Kumar et al. (2022) used SEM for the characterization of the fibre lumen collapsibility due to different refining energy. A qualitative analysis of the cross-section based on SEM images was published by Frazier et al. (2022) to evaluate the impact of dust particles on the fibre network. Morais et al. (2021) showed the densification effect of microfibrillated cellulose (MFC) as an additive in tissue paper using cross-sectional SEM images. Despite the frequent use of the SEM method in previous works, the application is often limited to qualitative analysis because the field of view (FoV) is small relative to the effort required in sample preparation.
Another approach to characterize 2D cross-sections of paper is based on optical image acquisition of paper samples embedded in resin, presented by Wiltsche et al. (2011). Their automated serial sectioning technique, which combines a rotary microtome and an optical microscope, allows them to examine the cross-section of papers and to analyze, for instance, the fibre wall thickness or the thickness of the coating layer. In this work this microtome technique is used for tissue cross-section evaluation for the first time. A detailed overview of the procedure is presented in the methods section.
A technique for tissue characterization that is not based on obtaining cross-sections is twin laser profilometry (TLP), which was introduced and applied by Sung et al. (2005) and Keller et al. (2009, 2022). TLP utilizes laser triangulation to measure the relative distance between both tissue surfaces and repeats this in the lateral direction to produce a map of the thickness distribution of a given area (Sung et al. 2005). The technique is also suitable for intrinsic thickness measurement of highly textured and embossed tissue, as out-of-plane deviations can be eliminated.
An effective but also elaborate approach to measure 3D bulk of fibre-based structures is based on X-ray microtomography (\(\mu\)-CT). This non-destructive method enables access to the inner bulk network including fibres and pores on a micrometer scale based on radiographs. The fibre network can be fully reconstructed in three dimensions, segmented and evaluated voxel-wise. \(\mu\)-CT measurements are performed in all fields of science, including the study of cellulose based networks such as paper and tissue. One of the first approaches of imaging the paper bulk structure was published by Samuelsen et al. (2001) based on synchrotron \(\mu\)-CT. Previous works relied predominantly on X-ray synchrotron radiation, which enables a high flux of photons, short measurement times and high resolution. The necessary equipment is expensive and located in specialized institutes such as the European Synchrotron Radiation Facility (ESRF). du Roscoat et al. (2005) have carried out extensive work on fibre and filler segmentation to evaluate pore size and filler content in printing and packaging paper, based on \(\mu\)-CT data from the ESRF. Another approach to characterize the pore structure of cardboard using the ESRF beam line was performed by Axelsson and Svensson (2010). Several other \(\mu\)-CT studies were carried out for printing or packaging papers. Focusing on their porosity (Machado Charry et al. 2018; Neumann et al. 2021) or connectivity properties like fibre-fibre bonds (Wernersson et al. 2014; Borodulina et al. 2016).
With respect to tissue paper, measurements and evaluations based on \(\mu\)-CT have rarely been described in previous works. Ismail et al. (2020) used \(\mu\)-CT for qualitative assessment of the crepe structure of the tissue surface together with SEM measurement. Keller et al. (2010) used a synchrotron device at the ESRF to image the z-direction of mass of paper towels. In a more recent study Keller et al. (2022) used \(\mu\)-CT data as basis for numerical simulation of the wetting behaviour of embossed tissue grades (DCT, TAD).
As already mentioned, the bulk structure of tissue paper is not comparable with other paper grades in terms of porosity, topography and thickness. Therefore, the evaluation of the bulk structure of this very open fibrous network needs to be adapted. First, the FoV must be significantly increased, especially for highly structured tissue such as TAD to obtain multiple repetitions of each structure. Secondly, the definition of the tissue bulk boundaries is difficult due to holes, protruding fibres and superimposed structures. Available algorithms from conventional paper imaging analysis, like the rolling ball approach used by Machado Charry et al. (2018); Neumann et al. (2021), are difficult to implement for tissue due to these holes and protruding fibres. Therefore, a morphological approach as applied by Borodulina et al. (2016) seems more appropriate for tissue. Keller et al. (2022) defined bulk boundaries by connecting z-direction extrema points at each in-plane position followed by a smoothing 3D moving average algorithm to account for protruding fibres.
In this work we have investigated the bulk structure of TAD tissue using different measurement methods and evaluated intrinsic bulk parameters such as thickness and porosity. Prior to bulk measurement we determined the apparent lateral structure size of the fabric patterns with an areal surface measurement system based on focus variation (Reitbauer et al. 2021). We analyzed the tissue samples using a destructive 2D method (microtome) and a non-invasive 3D measurement (\(\mu\)-CT). The 2D and 3D data are evaluated based on intrinsic thickness and porosity, and new parameters are also introduced to evaluate structural features. Besides discussing possible evaluation methods, this work also aims at discussing the challenges in tissue bulk analysis based on the obtained 2D and 3D data.
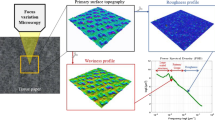
Surface topography of all four measured samples. K1 and K2 are kitchen towels, T1 and T2 toilet tissue. Surface measurements were carried out with the Infinite focus device from Alicona. Sq represents the areal quadratic mean square height of the filtered waviness surface and Lps the largest periodical structure size of the surface pattern
Materials and methods
Materials
In this study, four single-ply and unconverted TAD tissue samples were investigated with respect to their bulk and topographical properties. Sample K1 and K2 are kitchen towel grades and sample T1 and T2 refer to toilet paper grades. All tissue samples consist of a blend of softwood and hardwood kraft pulp fibres. The fibre properties, evaluated with a commercial flow cell based fibre analyzer (L&W fibretester), are listed in Table 1. The fines content evaluation is based on the ISO 160625-2 standard and the specified fibre length, width and fines content are length-weighted. To determine the single fibre properties the tissue samples were disintegrated. Basic properties of tissue paper samples like caliper (DIN EN ISO 12625-3) and basis weight (DIN EN ISO 12625-6) are determined based on a 10-ply stack. The values are listed in Table 2.
Surface topography
The topography of TAD tissue is mainly affected by the used TAD fabric. In order to classify our samples, measurement methods are necessary that provide suitable parameters to characterize the tissue topography. We used the principle of focus variation to determine the 3D surface data of the Yankee-side of the four samples. The measurement procedure of focus variation is explained in detail in previous work (Reitbauer et al. 2021) using the Infinite focus measurement device (IFM Model G3, Bruker-Alicona, Graz, Austria) to determine the mean structural size of the fabric patterns and their height. The tissue topography can be determined by the variation of the focal point within the z-direction. The height distance between the focal point of tissue surface and the optical sensor is measured. By repeating this procedure for the entire lateral size of the sample, a continuous surface profile can be obtained. In post processing a waviness filter was applied to eliminate small structures like single fibres. Consequently, only large structures like the TAD fabric pattern are visible. The 3D surface topography for all samples is shown in Fig. 1. As height parameter, the areal based root-mean-square height (Sq), also noted for each sample in Fig. 1, was used. To determine the size of the apparent periodical structures a power spectral density (PSD) analysis was carried out. Based on the PSD analysis, which is described in detail in Reitbauer et al. (2021), the largest periodical structure size (Lps) can be calculated and is shown in Fig. 1. Based on the Lps, the necessary FoV for subsequent measurements such as \(\mu\)-CT can be defined for each sample. We set the minimum length of the FoV in \(\mu\)-CT measurements to 3.85 mm to depict at least twice the size of one fabric structure.
Image processing sequence of microtome data. The grey value raw image slices a of each sample were binarized with the method of Otsu (Otsu et al. 1979). b shows the cross-section of the segmented tissue samples. Tissue bulk volume and boundaries are determined with four different expansion diameters. c shows the bulk with an expansion diameter of 15 μm. d shows the detection of the center line (zoomed in view)
Sample configuration for the \(\mu\)-CT measurement. The schematic sample set up within the Kapton tube in x–y and x–z direction is depicted. Three tissue strips from one sample are separated and immobilized with Kapton strips (orange) based on the work of (Neumann et al. 2021). The inner diameter of the Kapton tube is 2 mm and the minimum length of the scan in z-direction is 3.85 mm
Microtome measurement and evaluation procedure
Measurement and sample configuration
As already mentioned in the introduction, in previous works tissue bulk characterization has often been based on 2D data of the cross-section. In contrast to SEM measurements to determine tissue cross-section, microtome measurement does not require special preparation requirements in addition to embedding. Furthermore, a much larger FoV can be obtained at a given resolution. The instrumental set-up used in this study is based on the development of Wiltsche et al. (2011), although minor adjustments were necessary for tissue paper. One adaptation, for instance, is the increase of the FoV to get a robust amount of periodical structures, such as fabric patterns and crepe, captured. The cross-section is observed with a moveable optical microscope (ALC1 3-CMOS, Bruker-Alicona, Graz, Austria). By moving the microscope, a series of images, which are stitched together in a post-processing step, is captured. This enables a large FoV combined with a high resolution. The used magnification was 10\(\times\), resulting in a pixel size of 0.614 μm at a stitched cross section length of 18 mm per cut. A sufficient amount of periodic patterns from the TAD fabric is given when considering the Lps of the observed samples (see Fig. 1). Each sample was cut both in MD and CD and embedded with a methacrylate-based resin into the specimen holder. The resin immobilizes the tissue and allows smooth cuts in the subsequent measurement. Herein, a microtome together with a diamond knife is used to cut slices off the embedded sample. In total four cuts per direction and sample were acquired at a distance between the cuts of 1 mm, which results in eight cross-section profiles with 18 mm length per sample.
Segmentation of fibre network
Image processing of microtome data was performed using MATLAB (R2019b - MathWorks, Massachusetts, USA). The procedure is depicted in Fig. 2. Based on the segmentation method of Otsu (see Otsu et al. 1979), the raw image slices (a) were binarized (b) with an adaptive threshold for each tissue sample based on the background mean grey value. The subsequent steps (c) and (d) are discussed in the results section.
Overview of the fibre segmentation of the reconstructed \(\mu\)-CT data. a shows a raw image slice from the prior \(\mu\)-CT measurement and reconstruction. For the segmentation of fibres, air and Kapton a deep learning approach with the convolutional network U-Net was applied (Ronneberger et al. 2015). The final segmentation is shown in (b) and for the segmented fibres only in (c). The virtual volume of the segmentation is shown in (d)
\(\mu\)-CT measurement and evaluation procedure
Measurement and sample configuration
For 3D characterization we used a \(\mu\)-CT measurement system. Compared to synchrotron facilities, the measurement time is longer, however, resolution is high and access to such devices is much easier. Scans were performed using the lab-scale UniTOM HR device (TESCAN ORSAY HOLDING, Brno, Czech Republic). The acceleration X-ray voltage was 60 kV, and the target current was 91 μA. For each scan, 2286 projections were acquired with an angular range of 360 degrees. Each projection was taken with a DEXELA camera with an exposure of 1.29 s, five averages, and a two-times binning. The isotropic voxel size for all scans and samples was 1.2 μm. For each sample, five scans were stacked in the z-direction to have a FoV of about 1.83 mm \(\times\) 3.85 mm. This FoV is sufficient to capture the apparent periodic structures caused by the TAD fabric.
A stacked sample configuration, as proposed by Neumann et al. (2021), was used to increase the volume of tissue material per scan. A representation of this configuration is depicted in Fig. 3. Three layers of tissue were stacked in a Kapton tube with a 2 mm inner diameter and separated from each other with Kapton strips. The use of Kapton (Polyimide thin film) as a sample holder and fixation of the tissue during the scan is explained by its transparency for X-rays, moreover it doesn’t interfere with the absorption contrast (Neumann et al. 2021). The described measurement configuration allows the stacking of scans along the z-axis, making a larger sample volume accessible. In this work, five scans were stacked to create a vertical range at least twice the largest periodical structure size (Lps) (see Fig. 1). Based on the height of one scan, which depends on the set-up during the \(\mu\)-CT measurement, the final length along the z-axis is 3.85 mm. The tissue strips within the Kapton tube were cut with a laser cutter to prevent any changes in the fibre network due to mechanical treatment. Each sample stack consists of a cut in machine- (MD), cross- (CD) and transversal direction from different layers. After reconstruction of the scans with the Panthera (TESCAN XRE) software, the data of the scans are available for further processing as a sequence of several thousands 2D raw images.
Segmentation of fibre network
For all successive evaluations the Dragonfly Software (Version 2022.1-ORS Inc, Montreal, Canada) was used. A deep learning segmentation approach was used to separate the raw data into the available fractions of fibres, air, and Kapton. The segmentation procedure is depicted in Fig. 4a–d. The convolutional network chosen was originally used for biomedical image segmentation and is called U-Net (Ronneberger et al. 2015). As a starting point for the training of the network, single slices (as shown in Fig. 4a) of one dataset were segmented in fibre and non-fibre phase (air and Kapton) based on the method of Otsu (Otsu et al. 1979). After this initial training, at least seven arbitrary slices throughout the dataset were used to train the U-Net model for semantic segmentation. This enables a segmentation not only in xy-direction (2D) but also considers the z-direction (3D input dimension). The training of the U-Net model is divided in so called epochs. By means of a loss function, the progress of the training can be followed after each epoch. As a rule, the coefficient of the loss function decreases over the epoch number until no more changes occur. At this point the model can be considered trained and is applied to all slices in the sample dataset. The background contrast of each sample dataset was slightly different, so the the weighting parameters for the U-Net model were adapted for each tissue sample and then applied on the whole dataset. After segmentation, the fibre network of all four datasets is further optimized by removing outlier data and single fibres which are not connected to the main fibre network.
The transition phase (dark layer between fibres and air) is a common artefact from X-ray measurements. This transition phase can be assigned either to the fibre or air fraction. It is still the subject of current investigations whether this transition zone is rather air or rather fibre and any decision will result in general under- or overestimation of the fibre phase (Machado Charry et al. 2018). We assigned most of the transition phase to the air fraction, which probably leads to a slight overall underestimation of fibres.
Results and discussion
Tissue bulk characterization
After data acquisition and fibre network segmentation, both 2D and 3D datasets are used for tissue bulk characterization. Due to the low basis weight and the applied processes during production, such as moulding and creping, the bulk and surface structure of tissue in z-direction is hard to define. Single fibres protruding from the fibre surface and holes within the structure pose a challenge when it comes to the definition of the bulk and consequently the porosity. Looking into the intrinsic bulk properties of tissue samples and especially when comparing different samples it may very well be of interest whether different definitions of the bulk boundaries lead to different results. Therefore, we considered several bulk boundaries using the so called expansion diameters by expanding the diameter of a disk for 2D data and of a sphere for 3D data, which are also slightly different for 2D and 3D data since the resolution of both methods differ. Visual results are shown for some expansion diameters in Fig. 5a–c where 3D \(\mu\)-CT based data are represented as 2D cuts. With growing expansion sphere diameter, the considered bulk is increasing as well. The marked regions show two positions where the different expansion diameters have a significant impact. The area between fibres can be assigned as outside or inside of the bulk boundary and thus is either considered as a void or as part of the continuous fibre network in further evaluations. Intrinsic parameters such as thickness and porosity are obviously affected strongly, based on the affiliation of a region to bulk or to non-bulk. The question arises which expansion of the bulk is suitable and realistic for further considerations when comparing for instance the bulk for an expansion sphere diameter of 0.02 mm (see Fig. 5a) to 0.1 mm (c). Considering for example porosity and its effect on water uptake the void space within the bulk boundary might be overrepresented with the high expansion diameter, but it could also be underrepresented for smaller diameters. The intrinsic fibre network parameters thickness and porosity, discussed in the following, should allow a deeper understanding of the effect of bulk expansion applied to 2D and 3D data.
For the 2D microtome data the bulk of the segmented cross section images was evaluated based on a closed mask morphological approach, which includes dilatation of pixels followed by erosion (see Fig. 2c) (Said et al. 2016). Based on the chosen expansion diameters, the bulk boundaries vary. Diameters from 25 to 200 pixel corresponding to 15–120 μm were used. The definition of the center line, which is depicted in Fig. 2d, is based on the number of pixels between the edges in horizontal direction of the largest expansion diameter (200 pixel). The largest expansion diameters smooth the edges so that the effects of individual features such as protruding fibre on the center line detection are minimized. Treating the binarized images as a matrix of zeros (black) and ones (white), the number of white pixels (ones) between the edges is determined for each row, with the center pixel of each row constituting the center line. The center line is used to determine the actual length of the cut and for further evaluation like thickness variations.
The applied approach to determine the bulk boundaries for the 3D \(\mu\)-CT data is slightly different, as a morphological approach in three dimensions increases the computational effort significantly. Therefore, five expansion spheres with diameters from 17 to 100 μm were defined to fill contiguous pores with a diameter smaller than the corresponding value. Thus, different bulk volumes or bulk boundaries are taken into account depending on the distance of fibres to each other.
In general, 2D data based on the microtome is considered in pixel (pixel size: 0.617 μm) while 3D data based on the \(\mu\)-CT is expressed in voxel (voxel size: 1.2 μm).
Intrinsic thickness
In contrast to the caliper, the intrinsic thickness (μm) of tissue is difficult to determine due to the superimposed structures such as patterns and crepe. The different bulk expansion diameters of course bias the thickness measurement. For 2D microtome data, the thickness is determined by calculating the horizontal distance between the edges for each row of the given bulk expansion (see Fig. 2d). Since we are comparing the intrinsic properties to standard parameters, we specified that the intrinsic thickness is represented perpendicular to the x-y plane, similar to caliper measurements. The thickness distribution along the length of the center line is then evaluated and plotted in a cumulative probability curve for each sample. The average thickness value is based on the mean of the thickness distribution. The cumulated thickness distribution of sample K2 is shown in Fig. 6a. The thickness for the 3D \(\mu\)-CT data is evaluated based on the distribution of the thickness mesh used. The thickness is calculated as the diameter of a hypothetical sphere that fits into each boundary point of the mesh, where the mesh size can be adjusted based on the given feature size. Here, we defined a mesh size of 2 voxels in the x, y, and z directions to sufficiently account for individual fibres and thickness variances as well. The mesh encloses the edges of the tissue mass and allows accurate spatial determination of thickness at each position (see Fig. 5d). Similar to the 2D evaluation, 3D based intrinsic thickness is represented in a cumulative probability curve (see Fig. 6b), while the average thickness value is based on the mean of the distribution.
Figure 6 compares the effect of increasing expansion diameter on the intrinsic thickness distribution as an example for sample K2 for both 2D and 3D evaluation. For small expansion diameters (blue), the slope of the probability curves indicates a narrow thickness distribution that becomes increasingly flat with increasing expansion diameter (red). The bulk enclosed with expansion diameters such as 15 and 17 μm is discontinuous and can be described as ragged, while high expansion diameters connect more and more network segments to one continuous bulk structure, increasing the intrinsic thickness and its variability in general. The 2D based microtome data (a) tends to show lower thickness values than the 3D \(\mu\)-CT data (b). At a cumulative probability of 50 % and an expansion diameter of 30 μm the 2D data of sample K2 shows a thickness of 31 μm and the 3D data shows a thickness of 43 μm. The difference between both methods is increasing with larger expansion diameter. When comparing the highest expansion diameters (red) at 50 % cumulated probability the difference in thickness increases up to 30 μm. In addition, a broader distribution for higher expansion diameters can be seen in the 3D data (b) compared to the 2D data (a). These thickness differences may be due to the different expansion algorithms resulting from the 2D and 3D observations. In the 2D microtome data, a disk with specific diameters is used for bulk determination, however, expansion can be only considered in the two dimensional cross section, whereas in the 3D data, a sphere connects the fibres in all three spatial directions. This can lead to increased thickness values, as there is simply more segmented fibre material available during expansion.
The mean thickness values of all samples are compared in Fig. 7. The mean thickness values are based on the thickness distribution of each sample and are expressed over the bulk expansion diameter for the 2D microtome data in Fig. 7 as dashed lines and for the 3D \(\mu\)-CT data as solid lines. For both methods sample T2 shows the highest mean thickness values, at lowest expansion. However, above 40 μm expansion diameters the thickness spreads between 2D and 3D based data. In general, all samples show higher thickness values and a steeper curve with higher expansion diameters for the 3D based evaluation compared to the 2D cross-sectional evaluation. Similar to T2, sample K2 also shows the same mean thickness at lowest expansion diameter slightly over 30 μm and spreads with increasing expansion diameter between 2D and 3D data. Sample K1 and T1 show a similar trend with increasing expansion diameter for both 2D and 3D data, although the off-set is different. In contrast to the 3D evaluation, K2 does not show the lowest mean thickness based on the 2D data at the lowest expansion diameter, but only for higher expansion diameter. Thus, when comparing different tissue samples with respect to their intrinsic thickness the selected expansion diameter for the fibre network boundary determination has a great influence the obtained results. Other bulk boundary detecting algorithms, such as the rolling ball approach presented in Machado Charry et al. (2018), are also able to vary the size of the used sphere, but the loose network structure of tissue containing multiple holes may cause problems. We propose to consider multiple bulk expansions using varying expansion diameters for the characterization of intrinsic thickness and associated properties for tissue to avoid a possibly misleading evaluation. The results are strongly influenced by the number and intensity of surface and bulk features such as protruding fibres and dense/non-dense bulk regions.
After the determination and discussion of the intrinsic thickness the relation of these results to standard measurements for tissue such as the caliper and basis weight is of interest. The corresponding values are listed in Table 2. Caliper measurements are carried out with a micrometer, hence the absolute height in z-direction of a tissue paper at a given pressure is measured. A relation to the intrinsic thickness of both, 2D and 3D evaluations can not be observed. Especially, when considering the comparatively low caliper of sample T2 with 211 μm and the trend of the intrinsic thickness in Fig. 7, which shows by far the highest values. On the other hand, sample K2 has a caliper of 287 μm but shows the lowest intrinsic thickness values. Using the basis weight as additional variable, the apparent bulk based on caliper and bulk based on intrinsic thickness can be compared. Therefore, the mean intrinsic thickness of both the 2D and 3D evaluations with an expansion diameter of 30 μm is used to calculate the tissue bulk, since a comparable measurement point with the same expansion diameter is provided for both evaluations. These values, along with the caliper-based bulk values, are listed in Table 3. Besides the much lower values for the bulk based on 2D and 3D data, the trend of the samples is also almost reversed compared to the bulk calculated based on standard caliper. Sample T2 has the lowest bulk based on caliper with 9.13 cm3/g, but shows high values for the 2D and 3D based analysis. A similar, but opposing trend is also observed for sample K2 with a high value for caliper based bulk, but the lowest ones based on 2D and 3D intrinsic thickness. Figure 1 shows the root mean square heights (Sq) for each sample based on surface topography. Sample K2 shows the highest Sq value (68.95 μm) and sample T2 shows the lowest Sq (37.36 μm), indicating that the superimposed fabric structure greatly affects the caliper measurement and thus the bulk. Intrinsic thickness, on the other hand, is apparently unrelated to caliper measurements, at least for highly structured TAD tissues such as those analyzed in this work. An appropriate method based on the evaluation of the cross-section or mapping of the tissue surface is inevitable when it comes to the determination of the actual bulk and thickness of the base tissue paper.
Porosity and network connectivity
Porosity
In addition to allowing the determination of intrinsic thickness, both microtome and CT measurements provide detailed insight into fibre network properties, such as porosity. Porosity is defined as the ratio of the void space to the enveloping bulk in %. For 2D microtome evaluation, the area of pixels (\(mm^{2}\)) assigned to fibres and the total area based on bulk expansion are considered. With increasing expansion diameter the porosity of course also increases significantly. In the 3D \(\mu\)-CT evaluation of porosity, the volume (mm3) of the segmented fibre material and the volume of the enclosing bulk expansion are considered. The segmented data for 2D and 3D evaluation shows that the pore space within the tissue bulk is likely a continuous phase with small individual isolated pores in all measured samples. A closed pore network is not detectable, due to the open fibre network structure. The representation of the pore size distribution, as it is common for other paper grades (Machado Charry et al. 2018; Neumann et al. 2021), is therefore not meaningful.
Figure 8 depicts the porosity over the bulk expansion diameter for both 2D and 3D evaluation. At the lowest expansion diameter the 3D \(\mu\)-CT data (solid lines) show porosity values that are close to each other in a range from 44 % (T2) to 47.5 % (K2), with the values remaining similar at high expansion diameters, except for sample K2, which shows much higher values. This may indicate that with increasing expansion diameter more void volume is connected compared to the other samples, suggesting a more “ragged” fibre network structure and more protruding fibres in sample K2. In contrast to the 3D data, the 2D microtome data (dashed lines) show a broader porosity range from 45.5 % (K2) to 56.6 % (T2) at the lowest expansion diameter. The range remains broader also with increasing expansion diameters. Interestingly, the porosity results based on 2D and 3D evaluation do not show any relationship, especially when considering K2 and T2. K2 generally shows the lowest porosity values for the 2D data, while they are the highest for the 3D data and vice versa for sample T2. A possible reason might be the difference in the accessible respectively obtainable fibre structure between the 2D and 3D evaluation, since the \(\mu\)-CT based 3D evaluation is capable to reconstruct the whole tissue bulk structure in all spatial directions. For each tissue sample, at least eight slices were used for 2D evaluation, but a more comprehensive fibre network structure, as obtained with the \(\mu\)-CT data, cannot be represented. In addition, the 2D based microtome evaluation, together with other optical based cross-section segmentation methods, mostly detects fibres which are directed perpendicular or at least close to perpendicular to the image plane. Hence, fibres which are directed in an angle close to the image plane are not detected well (see Wiltsche et al. 2011) thus leading to a general underestimation of fibre network density, which is evident when comparing the absolute values of porosity between 2D and 3D evaluation in Fig. 8. Although, the 3D evaluated fibre network is also underestimated, since the mentioned transition phase is assigned to the air fraction. The microtome measurements were carried out in MD and CD to consider the different fibre directions, but still the 3D and 2D evaluation differ greatly in terms of porosity.
Comparing the results of the porosity evaluation in Fig. 8 and the intrinsic thickness evaluation in Fig. 7, no relationship is evident, especially for the 3D data. To further investigate this fact and the differences between the 3D and 2D evaluation in terms of porosity, a closer look at the fibre network conformation may be helpful. Therefore, we introduce a new parameter describing the fibre volume to fibre surface ratio Fv/Fs.
Fv/Fs—fibre network
This parameter represents the ratio of the fibre volume (3D)/area (2D) to the fibre surface (3D)/perimeter (2D). Unlike the other parameters the Fv/Fs is not affected by the bulk expansion, since fibre volume and surface remain the same for each tissue sample. By comparing the ratio of volume to surface area of the fibre phase, more information about the conformation of the fibre network is obtained. It will be affected by the bonded area in the structure and it should be able to differentiate between samples of comparable porosity for example, whether this porosity is caused by misleading fibre network boundaries due to protruding fibres while the rest of the network is rather dense in one case or by an overall loose network in the other. Besides the network conformation this parameter will also be affected by the fibre morphology in terms of coarseness. Coarser fibres will have a higher Fv/Fs ratio. The results for this analysis are shown in Fig. 9 for both, the 3D \(\mu\)-CT evaluation (a) and the 2D microtome evaluation (b). Similar to the porosity evaluation in Fig. 8 also the Fv/Fs parameter shows an opposite trend between 2D and 3D data. A comparison between the Fv/Fs parameter and the single fibre properties listed in Table 1 is of interest since the ratio will be affected by fibre coarseness. The fibres used for the kitchen towels K1 and K2 have a higher fibre length, fibre width and coarseness compared to the fibres used for the toilet tissues T1 and T2. Comparing the 2D based Fv/Fs parameter in Fig. 9b with the coarseness values, some interrelation can be observed for K1 and K2 which have a high coarseness and a high Fv/Fs ratio, while T1 and T2 have lower values for both parameters. This suggests that the 2D based evaluation depends more on single fibre properties than on the properties of the fibre network, which is most probably related to the fact that mainly fibres perpendicular or close to perpendicular to the cut plane will enter the evaluation, while fibres in the cut plane remain unnoticed. Therefore the bonded areas within the network may be underestimated and the coarseness of the detected fibres is the main driver for the results in case of 2D evaluation. In contrast, the 3D \(\mu\)-CT data in Fig. 9a show no correlation with coarseness. Instead, the high values for the Fv/Fs parameter for T1 and T2 most probably describe a denser network structure due to the generally smaller single fibre dimensions and a higher fines content. With a possible higher bonding and cross-linking probability the measured fibre surface (Fs) is reduced compared to the measured fibre volume (Fv), thus leading to a higher Fv/Fs ratio. The question of the earlier discussed missing correlation between porosity and intrinsic thickness remains. One possible explanation could be the irregular apparent dense and loose areas within the tissue, as they are not represented by the Fv/Fs ratio. Yet, the Fv/Fs parameter, together with the information about the individual fibres, provides a holistic view of the fibre network properties.
In summary, a direct comparison of 2D and 3D data in terms of network properties such as porosity is difficult, because the evaluation of the underlying data is different. Despite their evaluation differences both methods provide relevant information about intrinsic tissue properties, especially in terms of quantifying intrinsic thickness. Nevertheless, it must be emphasized once again that the correct evaluation of the given data, as performed by evaluating the effects of different expansion diameters on the intrinsic properties, is crucial for this purpose.
Conclusion and outlook
With the presented measurement techniques based on 2D microtome and 3D \(\mu\)-CT evaluation, the intrinsic properties of tissue paper such as thickness and porosity can be detected, compared and evaluated in a novel way. The variable expansion diameter algorithm used for both 2D and 3D data enables an overview of the effects of different bulk boundaries on intrinsic parameters such as thickness and porosity. It shows the importance of considering multiple fibre network boundaries for complex bulk structures, such as tissue paper, with their overlapping structures for further evaluation. This method might also be applicable for the characterization of other loose network structures like non-wovens. The focus of this work is on the comparison of 2D and 3D structural data of TAD tissues, the development of quantifiable evaluation routines, and the interpretation of the results. In terms of the evaluated intrinsic thickness both methods show similar trends for the observed tissue samples, whereas 2D based results tend to be lower with increasing bulk expansion diameter. Compared with standard caliper measurements both 2D and 3D evaluation based intrinsic thickness results show no relation to the measured caliper for highly structured TAD tissues used in this study. Also the apparent bulk based on the caliper measurement is compared with the bulk based on intrinsic thickness, showing no correlations. This fact might be interesting when it comes to the characterization of water absorption and the optimization of liquid transport through a tissue network. Both applied methods also allow the assessment of intrinsic network properties such as porosity. Together with the introduced fibre volume to fibre surface ratio (Fv/Fs) differences between 2D and 3D evaluation are evident. The 2D microtome data tends to be influenced by the single fibre properties, whereas 3D \(\mu\)-CT considers more the dependence on fibre connectivity and bonding properties. Therefore a \(\mu\)-CT-based representation of the 3D fibre network as skeletonized data might provide additional information about the connectivity and its dependency of fibre properties in future work. Another possible next step is to quantify the ratio of very dense to loose regions relative to the total bulk to determine the effects of processes like wet molding during production on the structure of the tissue network. Summarized, the present work highlights differences and procedures for the evaluation of the intrinsic properties of tissue paper based on a two-dimensional and a three-dimensional approach. The described procedures can provide substantial insights into loose and superimposed fibre network structures.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Commercial software was used.
References
Agarwal S, Srivastava P, Green SI, Phani AS (2023) The influence of microstructure on the tensile properties of a creped tissue paper: modeling and experiments. Mech Mater 176:104505. https://doi.org/10.1016/j.mechmat.2022.104505
Axelsson M, Svensson S (2010) 3D pore structure characterisation of paper. Pattern Anal Appl 13(2):159–172. https://doi.org/10.1007/s10044-009-0146-1
Borodulina S, Wernersson EL, Kulachenko A, Hendriks CL (2016) Extracting fiber and network connectivity data using microtomography images of paper. Nord Pulp Pap Res J 31(3):469–478. https://doi.org/10.3183/npprj-2016-31-03-p469-478
Das R, Pan K, Green S, Phani AS (2020) Creped tissue paper: a microarchitected fibrous network. Adv Eng Mater 2000777:1–6. https://doi.org/10.1002/adem.202000777
de Assis T, Pawlak J, Pal L, Jameel H, Venditti R, Reisinger LW, Kavalew D, Gonzalez RW (2019) Comparison of wood and non-wood market pulps for tissue paper application. BioResources 14(3):8781–8810. https://doi.org/10.15376/biores.14.3.6781-6810
de Assis T, Pawlak J, Pal L, Jameel H, Reisinger LW, Kavalew D, Campbell C, Pawlowska L, Gonzalez RW (2020) Comparison between uncreped and creped handsheets on tissue paper properties using a creping simulator unit. Cellulose 27(10):5981–5999. https://doi.org/10.1007/s10570-020-03163-0
du Roscoat SR, Bloch JF, Thibault X (2005) Characterisation of the 3D paper structure with X-Ray synchrotron radiation microtomography. In: 13th fundamental research symposium, Cambridge, 2005, pp 901–920. https://doi.org/10.15376/frc.2005.2.901
Frazier R, Zambrano F, Pawlak JJ, Peszlen I, Welsford D, Gonzalez R (2022) The tissue dust analysis system: a new device and methodology to quantify dusting and linting propensity in hygiene tissue papers. Cellulose 29(15):8387–8407. https://doi.org/10.1007/s10570-022-04779-0
Ismail MY, Patanen M, Kauppinen S, Kosonen H, Ristolainen M, Hall SA, Liimatainen H (2020) Surface analysis of tissue paper using laser scanning confocal microscopy and micro-computed topography. Cellulose 27:8989–9003. https://doi.org/10.1007/s10570-020-03399-w
Keller DS, Branca D, Kwon O (2009) Spatial partitioning of the structural properties of tissue and towel grades. In: 14th fundamental research symposium, Oxford, vol 2009, pp 693–719. https://doi.org/10.15376/frc.2009.2.693
Keller DS, du Roscoat SR, Huang Y (2010) Mapping the Z-direction of mass of paper towel grades using X-ray microtomography. In: 2010 progress in paper physics seminar, Montreal
Keller DS, Abedsoltan H, Reid J, Yin L (2022) Mesoscale water wetting of low-density fibrous structures: experimentation and simulation. In: 17th fundamental research symposium, Cambridge, 2022
Kullander J, Nilsson L, Barbier C (2012) Evaluation of furnishes for tissue manufacturing; wet pressing. Nord Pulp Pap Res J 27(5):947–951. https://doi.org/10.3183/NPPRJ-2012-27-05-p947-951
Kumar R, Zambrano F, Peszlen I, Venditti R, Pawlak J, Jameel H, Gonzalez R (2022) High-performance sustainable tissue paper from agricultural residue: a case study on fique fibers from Colombia. Cellulose 29(12):6907–6924. https://doi.org/10.1007/s10570-022-04687-3
Machado Charry E, Neumann M, Lahti J, Schennach R, Schmidt V, Zojer K (2018) Pore space extraction and characterization of sack paper using \(\mu\)-CT. J Microsc 272(1):35–46. https://doi.org/10.1111/jmi.12730
McCabe W (2011) Tissue properties and manufacturing—forming and TAD fabric. In: TAPPI presentation 2011
Morais FP, Carta AM, Amaral ME, Curto JM (2021) An innovative computational strategy to optimize different furnish compositions of tissue materials using micro/nanofibrillated cellulose and biopolymer as additives. Polymers. https://doi.org/10.3390/polym13152397
Morais FP, Vieira JC, Mendes AO, Carta AM, Costa AP, Fiadeiro PT, Curto JM, Amaral ME (2022) Characterization of absorbency properties on tissue paper materials with and without deco and micro embossing patterns. Cellulose 29(1):541–555. https://doi.org/10.1007/s10570-021-04328-1
Neumann M, Machado Charry E, Baikova E, Hilger A, Hirn U, Schennach R, Manke I, Schmidt V, Zojer K (2021) Capturing centimeter-scale local variations in paper pore space via \(\mu\) -CT: a benchmark study using calendered paper. Microsc Microanal. https://doi.org/10.1017/S1431927621012563
Neumann M, Machado Charry E, Zojer K, Schmidt V (2021) On variability and interdependence of local porosity and local tortuosity in porous materials: a case study for sack paper. Methodol Comput Appl Probab 23(2):613–627. https://doi.org/10.1007/s11009-019-09761-1
Otsu N, Smith PL, Reid DB, Environment C, Palo L, Alto P, Smith PL (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern SMC 9(1):62–66
Pawlak JJ, Frazier R, Vera RE, Wang Y, Gonzalez R (2022) Review: the softness of hygiene tissue. BioResources 17(2):3509–3550. https://doi.org/10.15376/biores.17.2.Pawlak
Reitbauer J, Harrer F, Eckhart R, Bauer W (2021) Focus variation technology as a tool for tissue surface characterization. Cellulose 28(11):6813–6827. https://doi.org/10.1007/s10570-021-03953-0
Ronneberger O, Fischer P, Brox T (2015) UNet: convolutional networks for biomedical image segmentation. IEEE Access 9:16591–16603. https://doi.org/10.1109/ACCESS.2021.3053408. arxiv:1505.04597
Said KAM, Jambek AB, Sulaiman N (2016) A study of image processing using morphological opening and closing processes. Int. J. Control Theory Appl 9(31):15–21
Samuelsen EJ, Gregersen OW, Houen PJ, Helle T, Raven C, Snigirev A (2001) Three-dimensional imaging of paper by use of synchrotron x-ray microtomography. J Pulp Pap Sci 27(2):50–53
Stankovská M, Fišerová M, Gigac J, Opálená E (2020) Blending impact of hardwood pulps with softwood pulp on tissue paper properties. Wood Research 65(3):447–458
Sung Y, CH H, Kwon O, Lee H, DS K (2005) Applications of thickness and apparent density mapping by laser profilometry. In: 13th fundamental research symposium, Cambridge, 2005, pp 961–1007. https://doi.org/10.15376/frc.2005.2.961
van Assis T, Reisinger LW, Pal L, Pawlak J, Jameel H, Gonzalez RW (2018) Understanding the effect of machine technology and cellulosic fibers on tissue properties: a review. BioResources 13(2):4593–4629. https://doi.org/10.15376/biores.13.2.deassis
Vieira JC, Mendes AdO, Ribeiro ML, Vieira AC, Carta AM, Fiadeiro PT, Costa AP (2022) Embossing pressure effect on mechanical and softness properties of industrial base tissue papers with finite element method validation. Materials. https://doi.org/10.3390/ma15124324
Wernersson EL, Borodulina S, Kulachenko A, Borgefors G (2014) Characterisations of fibre networks in paper using micro computed tomography images. Nord Pulp Pap Res J 29(3):468–475. https://doi.org/10.3183/npprj-2014-29-03-p468-475
Wiltsche M, Donoser M, Kritzinger J, Bauer W (2011) Automated serial sectioning applied to 3D paper structure analysis. J Microsc 242(2):197–205. https://doi.org/10.1111/j.1365-2818.2010.03459.x
Zambrano F, Wang Y, Zwilling JD, Venditti R, Jameel H, Rojas O, Gonzalez R (2021) Micro- and nanofibrillated cellulose from virgin and recycled fibers: a comparative study of its effects on the properties of hygiene tissue paper. Carbohydr Polym 254:117430. https://doi.org/10.1016/j.carbpol.2020.117430
Acknowledgments
We acknowledge the support by our industrial partner Andritz AG and the Austrian Research Promotion Agency (FFG).
Funding
Open access funding provided by Graz University of Technology. This study was funded by the Austrian Research Promotion Agency (FFG).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study concept and design. Material preparation, data collection and analysis were performed by JR, EMC and CS. The first draft of the manuscript was written by JR and all authors commented on all versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This chapter does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Reitbauer, J., Machado Charry, E., Eckhart, R. et al. Bulk characterization of highly structured tissue paper based on 2D and 3D evaluation methods. Cellulose 30, 7923–7938 (2023). https://doi.org/10.1007/s10570-023-05314-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-023-05314-5