Abstract
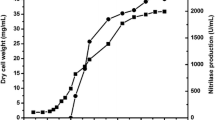
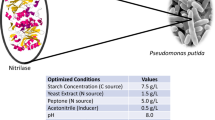
Nitrilases are important industrial enzymes to synthesize commercially useful acids from nitriles. In the present study, an aryl specific inducible nitrilase was produced from Alcaligenes faecalis MTCC 12629 which hydrolyzed 4-aminophenylacetonitrile to 4-aminophenylacetic acid. A bioprocess was developed to synthesize 4-aminophenylacetic acid from 4-aminophenylacetonitrile at 500 ml scale under optimized reaction conditions. The maximum activity of nitrilase for biotransformation of 4-aminophenylacetonitrile was recorded in 50 mM KH2PO4/K2HPO4 buffer of pH 7 at 45 °C. Operational stability of present enzyme was found at 45 °C for fed batch reaction. The bioprocess designed at 500 ml scale included 17 U/ml catalyst amount, 50 mM substrate addition after 30 min in total five feedings for 150 min at 45 °C. The approach of substrate feeding at lower amount resulted in accumulation of 175 mM product in 500 ml reaction after five feedings of substrate. The rate of product formation was found to be 2.5 g/gdcw/h with the volumetric productivity of 26.6 g/l.
Graphical Abstract





Similar content being viewed by others
References
Nigam KV, Arfi T, Kumar V, Shukla P (2017) Bioengineering of nitrilases towards its use as green catalyst: applications and perspectives. Indian J Microbiol 57:131–138
Bhalla TC, Kumar V, Kumar V, Thakur N (2018) Nitrile metabolizing enzymes in biocatalysis and biotransformation. Appl Biochem Biotechnol. https://doi.org/10.1007/s12010-018-2705-7
Gong JS, Lu ZM, Li H, Shi JS, Zhou ZM, Xu ZH (2012) Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Fact 11:142–148
Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409:258–266
Wohlgemuth R (2010) Biocatalysis - key to sustainable industrial chemistry. Curr Opin Biotech 21:713–724
Mukram I, Ramesh M, Monisha TR, Nayak AS, Karegoudar TB (2016) Biodegradation of butyronitrile and demonstration of its mineralization by Rhodococcus sp. MTB5. 3 Biotech 6:141
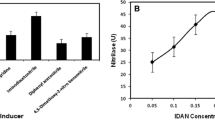
Nageshwar YVD, Sheelu G, Shambhu RR, Muluka H, Mehdi N, Malik MS, Kamal A (2011) Optimization of nitrilase production from Alcaligenes faecalis MTCC 10757 (IICT-A3): effect of inducers on substrate specificity. Bioproc Biosyst Eng 34:515–523
Zhu X, Gong J, Li H, Lu Z, Zhou Z, Shi J, Xu Z (2014) Screening, identification and culture optimization of a newly isolated aromatic nitrilase-producing bacterium–Pseudomonas putida CGMCC3830. Sheng Wu Gong Cheng Xue Bao 30:412–424
Kumar V, Bhalla TC (2013) Transformation of p-hydroxybenzonitrile to p-hydroxybenzoic acid using nitrilase activity of Gordonia terrae. Biocatal Biotransform 31:42–48
Vejvoda V, Sveda O, Kaplan O, Prikrylova V, Elisakova V, Himl M, Kubac D, Pelantova H, Kuzma M, Kren V, Martínkova L (2007) Biotransformation of heterocyclic dinitriles by Rhodococcus erythropolis and fungal nitrilases. Biotechnol Lett 29:1119–1124
Zhang ZJ, Xu JH, He YC, Ouyang LM, Liu YY, Imanaka T (2010) Efficient production of (R)-(-)-mandelic acid with highly substrate/product tolerant and enantioselective nitrilase of recombinant Alcaligenes sp. Process Biochem 45:887–891
Bhatia SK, Mehta PK, Bhatia RK, Bhalla TC (2013) Optimization of arylacetonitrilase production from Alcaligenes sp. MTCC 10675 and its application in mandelic acid synthesis. Appl Microbiol Biotechnol 98:83–94
Ya-Ping Xue, Jiao Biao, Hua Deng-En, Cheng Feng, Liu Zhi-Qiang, Zheng Yu-Guo (2017) Improving catalytic performance of an arylacetonitrilase by semirational engineering. Bioprocess Biosyst Eng 40:1565–1572
Martinkova L, Kren V (2018) Biocatalytic production of mandelic acid and analogues: a review and comparison with chemical processes. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-018-8894-8
Zhang XH, Liu ZQ, Xue YP, Wang YS, Yang B, Zheng YG (2018) Production of R-mandelic acid using nitrilase from recombinant E. coli cells immobilized with tris (hydroxymethyl) phosphine. Appl Biochem Biotechnol 184:1024–1035
Thakur N, Kumar V, Thakur S, Sharma N, Sheetal Bhalla TC (2018) Biotransformation of 4-hydroxyphenylacetonitrile to 4-hydroxyphenylacetic acid using whole cell arylacetonitrilases of Alcaligenes faecalis MTCC 12629. Process Biochem 73:117–123
Ekwuribe NN, Rodger Liddle (2011) Methods and compositions employing 4-aminophenylacetc acid compounds. United States Patent US 8,048,924 B2
Bedair AH, Ali FM, El-Agrody AM, Eid FA, El-Nassag MAA, El-Sherbeny G (2006) Preparation of 4-aminophenylacetic acid derivatives with promising antimicrobial activity. Acta Pharm 56:273–284
Egelkamp R, Schneider D, Hertel R, Daniel R (2017) Nitrile-degrading bacteria isolated from compost. Front Environ Sci. https://doi.org/10.3389/fenvs.2017.00056
Clouthier CM, Pelletier JN (2012) Expanding the organic toolbox: a guide to integrating biocatalysis in synthesis. Chem Soc Rev 41:1585–1605
Vergne-Vaxelaire C, Bordier FZ, Fossey A, Besnard-Gonnet M, Debard A, Mariage A et al (2013) Nitrilase activity screening on structurally diverse substrates: providing biocatalytic tools for organic synthesis. Adv Synth Catal 355:1763–1779
Fawcett JK, Scott JE (1960) A rapid and precise method for the determination of urea. J Clin Pathol 13:156–159
Wang H, Fan H, Sun H, Zhao L, Wei D (2015) Process development for the production of (R)-(−)-mandelic acid by recombinant Escherichia coli cells harboring nitrilase from Burkholderia cenocepacia J2315. Org Process Res Dev 19:2012–2016
Banerjee A, Kaul P, Banerjee U (2006) Purification and characterization of an enantioselective arylacetonitrilase from Pseudomonas putida. Arch Microbiol 184:407–418
Vesela AB, Krenkova A, Martinkova L (2015) Exploring the potential of fungal arylacetonitrilases in mandelic acid synthesis. Mol Biotechnol 57:466–474
Rustler S, Muller A, Windeisen V, Chmura A, Fernandes BCM, Kiziak C, Stolz A (2007) Conversion of mandelonitrile and phenylglycinenitrile by recombinant E. coli cells synthesizing a nitrilase from Pseudomonas fluorescens EBC191. Enzyme Microb Technol 40:598–606
He YC, Xu JH, Su JH, Zhou L (2010) Bioproduction of glycolic acid from glycolonitrile with a new bacterial isolate of Alcaligenes sp. ECU0401. Appl Biochem Biotechnol 160:1428–1440
Zhang XH, Liu ZQ, Xue YP, Xu M, Zheng YG (2016) Nirilase-catalyzed conversion of (R, S)-mandelonitrile by immobilized recombinant Escherichia coli cells harbouring nitrilase. Biotechnol Appl Biochem 63:479–489
Sun HH, Gao WY, Fan HY, Wang HL, Wei DZ (2015) Cloning, purification and evaluation of the enzymatic properties of a novel arylacetonitrilase from Luminiphilus syltensis NOR5-1B: a potential biocatalyst for the synthesis of mandelic acid and its derivatives. Biotechnol Lett 37:1655–1661
Bhatia SK, Mehta PK, Bhatia RK, Bhalla TC (2014) Optimization of arylacetonirilase production from Alcaligenes sp. MTCC 10675 and its application in mandelic acid synthesis. Appl Microbiol Biotechnol 98:83–94
Vesela AB, Petrickova A, Weyrauch P, Martinkova L (2013) Heterologous expression, purification and characterization of arylacetonitrilases from Nectria haematococca and Arthroderma benhamiae. Biocatal Biotransform 31:49–56
Nagasawa T, Nakamura T, Yamada H (1990) e-Caprolactam, a new powerful inducer for the formation of Rhodococcus rhodochrous J1 nitrilase. Arch Micobiol 155:13–17
Brenner C (2002) Catalysis in the nitrilase superfamily. Curr Opin Struct Biol 12:775–782
O’Reilly C, Turner PD (2003) The nitrilase family of CN hydrolysing enzymes – a comparative study. J Appl Microbiol 95:1161–1174
Yamamoto K, Fujimatsu I, Komatsu KI (1992) Purification and characterization of the nitrilase from Alcaligenes faecalis ATCC 8750 responsible for enantioselective hydrolysis of mandelonitrile. J Ferment Bioeng 73:425–430
Sosedov O, Baum S, Burger S, Matzer K, Kiziak C, Stolz A (2010) Construction and application of variants of the Pseudomonas fluorescens EBC191 arylacetonitrilase for increased production of acids or amides. Appl Environ Microbiol 76:3668–3674
Nagasawa T, Mauger J, Yamada H (2010) A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3-purification and characterization. Eur J Biochem 194:765–777
Layh N, Parratt J, Willetts A (1998) Characterization and partial purification of an enantioselective arylacetonitrilase from Pseudomonas fluorescens DSM 7155. J Mol Catal B Enzym 5:467–474
Zhu D, Mukherjee C, Yang Y, Rios BE, Gallagher DT, Smith NN, Biehl ER, Hua L (2008) A new nitrilase from Bradyrhizobium japonicum USDA 110 gene cloning, biochemical characterization and substrate specificity. J Biotechnol 133:327–333
Kaplan O, Vejvoda V, Charvatova-Pinvejcova A, Martínkova L (2006) Hyper induction of nitrilases in filamentous fungi. J Ind Microbiol Biotechnol 33:891–896
Prasad S, Misra A, Jangir VP, Awasthi A, Raj J, Bhalla TC (2006) A propionitrile-induced nitrilase of Rhodococcus sp. NDB 1165 and its application in nicotinic acid synthesis. World J Microbiol Biotechnol 23:345–353
Sharma NN, Sharma M, Kumar H, Bhalla TC (2006) Nocardia globerula NHB-2: bench scale production of nicotinic acid. Proc Biochem 41:2078–2081
Raj J, Singh N, Prasad S, Seth A, Bhalla TC (2007) Bioconversion of benzonitrile to benzoic acid using free and agar entrapped cells of Nocardia globerula NHB-2. Acta Microbiol Immunol Hung 54:79–88
Zhang ZJ, Xu JH, He YC, Ouyang LM, Liu YY (2011) Cloning and biochemical properties of a highly thermostable and enantioselective nitrilase from Alcaligenes sp. ECU0401 and its potential for (R)-(−)-mandelic acid production. Bioproc Biosyst Eng 34:315–322
Fan H, Chen L, Sun H, Wang H, Liu Q, Wei YRD (2017) Development of nitrilase-mediated process for phenylacetic acid production from phenylacetonitrile. Chem Pap 1985-1992
Chen J, Zheng YG, Shen YC (2010) Biotransformation of p-methoxyphenylacetonitrile into p-methoxyphenylacetic acid by resting cells of Bacillus subtilis. Biotech and Appl Biochem. https://doi.org/10.1042/BA20070106
Acknowledgement
The authors acknowledge University Grants Commission (UGC) New Delhi, India, for financial support in the form of UGC SAP SRF to Ms. Neerja Thakur and Faculty Fellowship to Prof. T. C. Bhalla under UGC-BSR scheme.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Thakur, N., Sharma, N.K., Thakur, S. et al. Bioprocess Development for the Synthesis of 4-Aminophenylacetic Acid Using Nitrilase Activity of Whole Cells of Alcaligenes faecalis MTCC 12629. Catal Lett 149, 2854–2863 (2019). https://doi.org/10.1007/s10562-019-02762-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10562-019-02762-2