Abstract
Three dimensional speckle tracking echocardiography (3D-STE) is a novel modality for the assessment of left ventricular strain (LVS). The aim of our study is to provide single vendor normative strain values measured with 3D-STE in healthy adult caucasians. One hundred fifty-five healthy subjects aged 20 to 72 years (≥ 28 subjects per decile) were prospectively included and examined with 2D and 3D transthoracic echocardiography. In 105 both 3D and 2D-STE were feasible (71%, mean age 44 ± 14 years, 51% female). Mean 3D tangential strain (3D-TS) was − 32 ± 2.9%. 3D global longitudinal strain (3D-GLS) demonstrated a significant but not very strong correlation with 2D-GLS values (− 19.7 ± 1.8% vs − 20.4 ± 2.2%, r = 0.462, p < 0.001). No gender difference was observed in 3D strain parameters. 3D-GLS decreased with increasing age stratum (p = 0.024). LVEF was associated with 3D-TS and 3D-GLS (r = − 0.819, p ≤ 0.001 for 3D-TS, p = − 0.477, r < 0.001, p = 0.001 for 3D-GLS). In this single vendor study age and gender-specific normative LV 3D-TS values were reported for healthy adult caucasians. In a significant proportion of the subjects 3D-STE was not feasible, but when feasible, 3D-STE shows excellent association with LVEF, and is therefore a promising novel modality for the assessment of the myocardial function, provided that issues of limited feasibility and temporal resolution are addressed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Speckle-tracking echocardiography (STE) is a novel imaging modality that allows quantitative assessment of global and segmental left ventricular (LV) myocardial function by measuring LV strain (LVS) in a manner largely independent of angle and ventricular geometry [1]. Myocardial deformation measurements allow early diagnosis of LV dysfunction by facilitating better risk stratification, reclassification and treatment in patients with cardiovascular disease [2]. 2D-STE, which is the established echocardiographic modality for LVS analysis, has the inherent limitations of planar simplification and apical foreshortening [3].
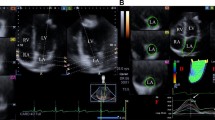
3D-STE has been developed with the aim of overcoming the technical limitations associated with 2D-STE. This technique allows tracking of speckle patterns that move out of the imaging plane. A novel myocardial deformation parameter, 3D tangential strain (3D-TS), which is defined as the greater of the 2 deformation values along the 2 diagonals of the quadrangular endocardial units formed by the analysis software (Fig. 1),could also be calculated with this modality [4]. Several studies, the largest being the NORRE study by the European Association of Cardiovascular Imaging (EACVI), have evaluated the LVS patterns by 3D-STE in healthy adults. Reference ranges in healthy adults were proposed, and age and gender-related distribution patterns of these parameters were described [5, 6].
The previous studies demonstrated significant differences in LVS values. These differences are attributed to the ultrasound (US) system [7], STE analysis software [8], and the population studied [5]. A recent meta-analysis demonstrated that both software vendor, US system and the population studied were independently associated with the variability in the LVS values measured by 3D-STE [6]. The NORRE study, the largest cohort of 3D-STE data on healthy individuals, used the Tomtec software (Tomtec, TomTec Imaging Systems, Unterschleissheim, Germany) for STE analysis. Two different US systems, Philips (Philips Medical Systems, Best, the Netherlands) and GE (GE Vingmed Ultrasound AS, Horten, Norway), were used for image acquisition in this study. The selected image acquisition and STE analysis system varies from study to study, and makes the comparability and interpretation of the results difficult. There is a need for population specific, single US system and single analysis software data on healthy individuals.
In the current study, our aim was threefold: (1) to provide normative values for 3D-STE in a cohort of healthy adult caucasians aged 20–72 years, obtained with a single-vendor US (Philips) and STE analysis platform (Tomtec) (2) to compare the feasibility and the distribution of the 2D and 3D-STE derived LVS; (3) and to evaluate the association of 3D-STE derived LVS patterns with baseline characteristics of the study participants.
Materials and methods
Study population
This prospective cohort study was conducted in 2014–2015 in 155 healthy volunteers aged 20 to 72 years, who were stratified into 5 age groups: 20 to 29, 30 to 39, 40 to 49, 50 to 59, and 60 to 72 years (n ≥ 28 in each group, equal distribution in sex). The subjects were recruited via an advertisement for healthy subjects. The inclusion criteria required that subjects had normal results on physical examination and electrocardiography (ECG). Subjects were excluded when they met any of the following criteria: prior cardiovascular disease; cardiovascular risk factors consisting of hypertension, diabetes mellitus or hypercholesterolemia; systemic disease or medication known to influence cardiac function; or the finding of cardiac abnormalities during examination. Professional athletes, morbidly obese subjects (body mass index, BMI > 40 kg/m2), pregnant women and women with breast implants were also excluded.
Clinical assessment
The assessment consisted of physical examination, 12-lead ECG, and echocardiography. Physical examination included height, weight, blood pressure, saturation, and results of heart, lungs and abdominal findings.
Echocardiographic image acquisition
All echocardiographic studies were performed by 2 experienced sonographers. Two-dimensional greyscale harmonic images were obtained in the left lateral decubitus position using an iE33 or EPIQ7 ultrasound system (Philips Medical Systems, Best, the Netherlands) equipped with a transthoracic broadband X5-1 matrix transducer (composed of 3040 elements with 1–5 MHz). Standard apical 4-chamber (A4C), 2-chamber (A2C) and 3-chamber (A3C) views were obtained for STE at frame rates of ≥ 50 frames/sec. Real-time 3D-TTE was performed immediately after the 2D-TTE with the same ultrasound unit and transducer. A minimum 4-beat full volume dataset of the LV was acquired from the apical window during a single breath hold. The acquired images were transferred to the analysis software on a dedicated workstation. Conventional echocardiographic measurements including detailed left atrial mitral inflow and pulmonary vein inflow measurements were performed using the proprietary software of the echocardiography machine [9,10,11]. 2D-STE and 3D-STE analyses were performed using Image Arena version 4.6 (TomTec Imaging Systems, Unterschleissheim, Germany) by 2 investigators experienced in the field (FOM) and (DB). The data sets were blinded for the observers. The average peak endocardial strain values given by the software were exported to a spreadsheet program.
Conventional echocardiographic measurements
We used the current guidelines of the American Society of Echocardiography and EACVI for chamber quantification [12]. From the A4C, pulsed-wave Doppler examination was performed to obtain peak mitral inflow velocities at early (E) and late (A) diastole and E-wave deceleration time (DT). Tissue Doppler imaging (TDI) was performed to obtain myocardial tissue velocity at the septal mitral annulus at early diastole (E’). 2D LV end-diastolic volume (LVEDV), LV end-systolic volume (LVESV) and LVEF were derived with the biplane method of disks based on one cardiac cycle.
2D-STE strain analysis
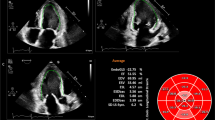
2D Cardiac Performance Analysis package of the Tomtec software was used for 2D-STE. The 2D-STE was defined as feasible when all the strain parameters were retrievable and the number of poorly tracking or non-tracking segments was ≤ 2. Cardiac cycles were defined by the position of the R-waves. End-diastole was tagged at the frame corresponding to peak of R wave. End-systole was tagged at the first frame at which LV has the minimal size following the reference R wave. The endocardial borders were manually traced in the A4C, A3C and A2C for the LS and in the mid-papillary level short-axis images for radial and circumferential strain. After the manual tracing, the software automatically tracked the endocardial border. Manual corrections were made when necessary. Peak systolic strain values were defined as the peak values on the curves during the ejection phase of one cardiac cycle. The 2D average global longitudinal strain, mid-papillary level average circumferential and radial strain (2D-MS-CS, 2D-MS-RS), LVEDV, LVESV and LVEF values were obtained.
3D-STE strain analysis
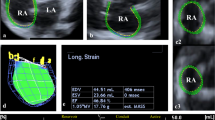
4D LV-analysis package of the Tomtec software was used for 3D-STE analysis. The software processed the 3D full volume dataset and produced the 2, 3 and 4 chamber views automatically. In the respective panels displaying these views, an axial line segment was aligned by the operator manually so that it was positioned between the center point of the mitral annulus and the LV apex. The software automatically traced the LV endocardium frame-by-frame in the next step. Manual corrections were made on the automatically traced endocardial borders as needed, in the end-systolic and end-diastolic frames. Finally, the software gave the peak and end-systolic 3D-TS, 3D-GLS, 3D-GCS, 3D-GRS, twist and torsion values as well as 3D-LVEDV, 3D-LVESV and 3D-LVEF. The analysis software derives the radial strain measurements from the endocardial area, according to the principle of volume conservation. Circumferential strain is derived from the endocardial border [13]. Tracking quality was classified as excellent, good or fair, and the signal to noise ratio was classified as good or bad, subjectively by the operator. 3D-STE analysis was defined as feasible when all the strain parameters were retrievable and the number of poorly tracking or non-tracking segments was ≤ 2 (Fig. 2) [4].
Statistical analysis
The data distribution was tested using histograms and the Shapiro–Wilk test. Continuous data were presented as mean ± standard deviation (SD). Categorical data was presented as frequencies and percentages. For comparison of normally distributed continuous between two groups with the Student’s t-test. In case of skewed distribution, the Mann–Whitney-U test was applied instead of t-test. For comparison of frequencies the χ2-test was used. For comparison of distribution of continuous variables between age strata, one-way ANOVA test was used. Analysis of correlation was performed with Pearson’s correlation coefficient. Multiple linear regression analysis was used for determining independent effect of predictors which were included in the prediction model. Two-tailed tests were used for correlation analysis and complete case analysis was the preferred method.
Intra-observer agreement was assessed by repeated analysis in a sample of 20 subjects (n = 4 per decile, randomly selected) > 1 month after the initial analysis, in randomly mixed order, and blinded to the initial results. Assessment of inter-observer agreement was performed by the second investigator (DB) in the same sample. The agreement between 2 measurements was determined by correlation analysis with scatter plots and Bland–Altman analysis with Bland–Altman plots for determination of intra and inter-observer variabilities. All statistical analyses were performed using SPSS statistics version 25 (IBM Corp., Armonk, NY, USA). A p value of less than 0.05 was considered statistically significant.
Results
Study population
Of the 155 subjects who came to our medical center, 147 were included in this study (age 45 ± 14 years, 50.3% female). Eight subjects were excluded due to limited echocardiographic acoustic windows (n = 2), valve pathology (n = 2), surgically closed patent ductus arteriosus (n = 1), hypertension (n = 1), morbid obesity (n = 1), and right bundle branch block on ECG (n = 1) [9]. Table 1 shows the characteristics of the study population.
Feasibility, temporal resolution and image quality
3D-STE was feasible in 117 of the 147 participants (79.6%) while 2D-STE was feasible in 130 participants (88.4%) (p < 0.001). When the patients in whom 3D-STE was feasible and non-feasible were compared, 3D-STE feasible participants had a greater BMI and a lower E-wave amplitude (Table 1). In 105 participants (71.4%) both 3D and 2D-STE were feasible and were included in the further analyses. Frame rates of the echocardiographic images were lower for 3D-STE compared with 2D-STE (30 ± 10 vs 60 ± 11 Hz, p < 0.001). Majority of the participants demonstrated good tracking quality (n = 58, 55.2%) and good signal to noise ratio (n = 78, 74.3%). Fourty-nine participants (47%) had at least one poorly tracked or non-tracked segments. The segments commonly associated with tracking problems were basal antero-septum, infero-septum and infero-lateral segments (Table 2).
LV strain
3D strain values are shown in Table 3, the distribution of the values in age and gender strata is shown in Fig. 3. Mean 3D-TS was − 32 ± 2.9% in the total study population. There were no significant gender-related differences with 3D-STE strain analysis. Among 3D strain parameters, only 3D-GLS decreased with increasing age strata (p = 0.024). 3D-LVEDV and 3D-LVESV were lower in female participants while 3D-LVEF did not differ between genders. The 3D-GLS values were lower compared with 2D-GLS values (− 19.7 ± 1.8% vs − 20.4 ± 2.2%, p = 0.001) (Fig. 4a). There was a statistically significant, but moderate correlation with a bias of − 0.7 ± 2.1 between 3D-GLS and 2D-GLS (r = 0.462, p < 0.001) (Fig. 4b, c).
a Distribution of 3D-TS, 3D-GLS, 2D-GLS in the study population b Scatter plot graph demonstrating correlation of 3D-GLS and 2D-GLS measurements c Bland–Altman graph demonstrating agreement between of 3D-GLS versus 2D-GLS measurements. Error bars represent 95% confidence interval of the mean, r Pearson’s r coefficient, TS tangential strain, GLS global longitudinal strain
Variables associated with 3D-TS and 3D-GLS
Results of the univariate and multivariate analyses are summarized in Table 4. In univariate analysis, lower end-diastolic intraventricular septum diameter (IVSd) (r = 0.229, p = 0.021), LVESV (r = − 0.419, p < 0.001) and LVEF (r = − 0.819, r < 0.001) were associated with 3D-TS. In a model consisting of age, systolic blood pressure, PWd, E wave, E’ wave amplitude and LVEF, only LVEF was independently associated with 3D-GLS (USβ = − 0.156, Sβ = − 0.346, R2 = 0.307, p = 0.001). Diastolic blood pressure demonstrated a strong correlation with systolic blood pressure, and E/A had a strong correlation with E wave amplitude. Thus, these 2 parameters were excluded from the model. Previous studies suggest that the effect of heart rate on myocardial strain is mainly via the changes in volumes and contractility. This parameter was also excluded from the model as a result. Multivariate analysis was not performed for 3D-TS, because the 3 predictors, IVSd, LVESV and LVEF correlated significantly with each other.
Intra-observer and inter-observer agreement
The intra-observer variability was − 1.1 ± 2.7% and the inter-observer variability was − 2.5 ± 3.1% for 3D-TS. The intra and inter-observer variabilities for 3D-GLS were − 0.9 ± 2% and − 1 ± 2.1% (Fig. 5).
Discussion
The current study presented normative values for 3D-STE strain variables for a healthy caucasian population. The feasibility of 3D-STE was lower than 2D-STE. The 3D-GLS values correlated significantly with 2D-GLS. LVEF was the only baseline characteristic that was associated with both 3D-TS and 3D-GLS.
3D-STE: normative data
The mean 3D-STE strain values were similar to published data on the same population [8], however lower than the multinational registries [5], despite similar BMI values. Normative ranges of 2D transthoracic echocardiography parameters were higher compared with the other populations in previous studies. This findings were explained with anthropometric characteristics, since Dutch people are the tallest of the world [9]. Larger body surface area with larger acoustic window might be expected to increase accuracy of the measurements. Conversely, larger lung tissue, larger BMI with resultant increase in subcutaneous adipose tissue might decrease the image quality and the accuracy of the strain measurements. Previously increased body size was found associated with decreased strain measurements using B-mode strain [14]. Population-specific genetic, structural or physiological characteristics other than anthropometric measures might also result in these differences.
The variability might alternatively result from the US-system or the STE analysis software. Gayat et al. demonstrated that US vendor (Toshiba versus Philips) was associated with variability in strain values [15]. Factors related with the US system such as probe specifications and image processing, and differences in temporal and spatial resolution of the system, as well as differences resulting from strain definitions (tangential vs area), region of interest selection (endocardial vs full thickness) and temporal definitions (ie. timing of the end-systole and end-diastole) are the potential sources of variability (Appendix 2) [16]. The EACVI-ASE Strain Standardization Task Force provides ongoing work in characterizing the inter-vendor variability and standardization of strain imaging with STE [17].
3D-STE versus 2D-STE
Along with the solution of geometric simplification and apical shortening, 3D-STE is equipped with the capability of producing novel 3D strain measures such as 3D-TS [4]. Nevertheless, limited temporal resolution is a well-known limitation of 3D-STE, when compared with 2D-STE. Recommended frame rate is 40 to 80 Hz for 3D-STE, although reported frame rates are usually lower [6]. Frame rates achieved with 3D-STE in our study were similar to the values obtained in previous studies but below the recommended range as well as the frame rates achieved by 2D-STE.
We compared measurement of GLS, the LV strain parameter with the most robust evidence, with 2D-STE and 3D-STE. 3D-GLS demonstrated significant correlation with 2D-GLS. The normative range of 3D-GLS was significantly lower than with 2D-GLS, which is consistent with the previous findings [18]. In a recent study apical foreshortening resulted in up to 23% overestimation in 2D-STE [3]. Lower 3D-GLS values compared with 2D-GLS in this study might be the result of a more accurate localization of the apex. We suggest that 3D-GLS could be implemented in measurement of GLS, but the validated reference ranges for 2D-GLS should not be used interchangeably with 3D-GLS.
Relationships of 3D-STE with age, sex and LVEF
Previous studies have found an age-related decrease in 3D-GLS [8, 19]; however, large scale studies and meta-analyses failed to confirm these patterns [20]. In our study, 3D-GLS decreased, while 3D-GCS and 3D-GRS did not change with increasing age. 3D-GLS was not associated, and the remaining 3D strain parameters were weakly associated with age in the recent NORRE study [5]. Different studies have shown a decreased [21], or increased [19] 3D-GCS and 3D-GRS values with increasing age.
LVS values has been found higher in women in previous studies [5]. This is usually attributed to higher LVEF in women [9]. However, a recent meta-analysis failed to demonstrate this difference [6]. The differences in 3D strain values were not statistically different between genders in this study. This finding paralleled similar 3D-LVEF values between genders. Lack of replicability in the age and gender-related changes among previous studies and also the current one has been attributed to the complex interplay between population-specific and vendor-specific factors. It is suggested that gender-related differences in LV strain was attributed to differences in body size between genders using B-mode strain measurements. Populations with closer body size ranges between genders might have closer LV strain values [14]. Breast tissue in women sometimes makes localization of the true apex difficult. Resulted foreshortening might also result in higher values in women [3].
Another possible factor underlying this variability is sample size. Especially for cohorts that consist of healthy individuals, distribution range of both strain and volumetric parameters is narrower. As a result, retrieving significant test results would require considerably large sample sizes in such studies. The statistically nonsignificant but higher 3D-TS and 3D-GLS and 3D-LVEF values in women, and lower 3D-TS, 3D-LVEDV and 3D-LVEF with increased age might be associated with these last 2 factors. The large international registries [5] as well as studies on populations with different ethnic background, will delineate ethnic as well as age and gender-related differences with regard to myocardial deformation patterns.
LVEF was the only factor associated with both 3D-TS and 3D-GLS. This finding is in accordance with the findings from other studies in healthy individuals [5]. When 3D-STE and 2D-STE are compared, 3D-TS and 3D-GLS correlated stronger with LVEF compared with 2D-GLS in previous studies [22]. GLS is a sensitive marker of LV global systolic function and might be impaired in subclinical cardiac damage, when LVEF is within normal limits [8]. However, in healthy individuals without cardiovascular disease, GLS is expected to correlate better with LVEF. We demonstrated that 3D-TS and 3D-GLS are sensitive to changes in LVEF values within normal range in healthy adults.
Feasibility and reproducibility
The patients for the current study were initially enrolled for 2D-STE. The feasibility of 3D-STE reported herein might reflect the real-world experience better as a result. Yet, the feasibility of 3D-STE in our study was higher than some of the reported values as low as 71% [23]. Higher BMI is associated with decreased transthoracic echocardiography image quality. Resultant decreased feasibilty was demonstrated in the current study. Lower E wave amplitude in the 3D-STE nonfeasible group might be associated with increased BMI [24].
The reproducibility of 3D-TS and 3D-GLS was acceptable [8]. The manual corrections performed during the endocardial tracing is an important source of variability for 3D-STE. Implementation of fully automated border tracking and machine-learning algorithms by the 3D-STE systems will potentially eliminate the problems related with reproducibility in the future [25].
Limitations of the study
The software platform used for 3D-STE only allows endocardial strain analysis. 3D-STE analysis package didn’t allow exclusion of non-tracking or poorly-tracking segments. An approach focusing on global average values was preferred as a result. Demonstration of the intervendor variability would have had incremental role in this study. However, we think that only inter-vendor agreement, but not the validity of a certain vendor, could be assessed in such a study, since there is no currently available noninvasive gold standard that the in-vivo accuracy of the available platforms could be tested against.
Conclusion
Age and sex-related changes as well as normative values were provided in this single US system, single analysis software study on healthy adult Caucasians. 3D-STE demonstrated reasonable feasibility and excellent agreement with LVEF. 3D-STE should be implemented in clinical evaluation of the LV function. The findings herein support interpretation of 3D-STE based myocardial deformation with vendor- and population-specific reference ranges. The 3D strain values should not be used and interpreted interchangeably with 2D strain values.
Data availability
The data is available and permission might be sought per reasonable request.
References
Voigt JU, Cvijic M (2019) 2- and 3-dimensional myocardial strain in cardiac health and disease. JACC Cardiovasc Imaging 12(9):1849–1863
Cikes M, Solomon SD (2015) Beyond ejection fraction: an integrative approach for assessment of cardiac structure and function in heart failure. Eur Heart J 37(21):1642–1650
Unlu S et al (2020) Impact of apical foreshortening on deformation measurements: a report from the EACVI-ASE Strain Standardization Task Force. Eur Heart J Cardiovasc Imaging 21(3):337–343
Pedrizzetti G et al (2014) Three-dimensional principal strain analysis for characterizing subclinical changes in left ventricular function. J Am Soc Echocardiogr 27(10):1041–1050
Bernard A et al (2017) 3D echocardiographic reference ranges for normal left ventricular volumes and strain: results from the EACVI NORRE study. Eur Heart J Cardiovasc Imaging 18(4):475–483
Truong VT et al (2019) Normal ranges of left ventricular strain by three-dimensional speckle-tracking echocardiography in adults: a systematic review and meta-analysis. J Am Soc Echocardiogr 32(12):1586–1597
Yuda S et al (2014) Inter-vendor variability of left ventricular volumes and strains determined by three-dimensional speckle tracking echocardiography. Echocardiography 31(5):597–604
Muraru D et al (2014) Left ventricular myocardial strain by three-dimensional speckle-tracking echocardiography in healthy subjects: reference values and analysis of their physiologic and technical determinants. J Am Soc Echocardiogr 27(8):858–871
van Grootel RWJ et al (2017) Echocardiographic chamber quantification in a healthy Dutch population. Netherlands Heart J 25(12):682–690
van Grootel RWJ et al (2018) In-depth echocardiographic analysis of left atrial function in healthy adults using speckle tracking echocardiography and volumetric analysis. Echocardiography 35(12):1956–1965
McGhie JS et al (2017) Quantitative assessment of the entire right ventricle from one acoustic window: an attractive approach. Eur Heart J Cardiovasc Imaging 18(7):754–762
Lang RM et al (2015) Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur Heart J Cardiovasc Imaging 16(3):233–270
Satriano A et al (2017) Clinical feasibility and validation of 3D principal strain analysis from cine MRI: comparison to 2D strain by MRI and 3D speckle tracking echocardiography. Int J Cardiovasc Imaging 33(12):1979–1992
Støylen A, Mølmen HE, Dalen H (2019) Left ventricular global strains by linear measurements in three dimensions: interrelations and relations to age, gender and body size in the HUNT Study. Open Heart 6(2):e001050
Gayat E et al (2011) Reproducibility and inter-vendor variability of left ventricular deformation measurements by three-dimensional speckle-tracking echocardiography. J Am Soc Echocardiogr 24(8):878–885
Amzulescu MS et al (2019) Myocardial strain imaging: review of general principles, validation, and sources of discrepancies. Eur Heart J Cardiovasc Imaging 20(6):605–619
Voigt JU et al (2015) Definitions for a common standard for 2D speckle tracking echocardiography: consensus document of the EACVI/ASE/Industry Task Force to standardize deformation imaging. J Am Soc Echocardiogr 28(2):183–193
Badano LP et al (2013) Use of three-dimensional speckle tracking to assess left ventricular myocardial mechanics: inter-vendor consistency and reproducibility of strain measurements. Eur Heart J Cardiovasc Imaging 14(3):285–293
Kleijn SA et al (2015) Normal reference values of left ventricular strain using three-dimensional speckle tracking echocardiography: results from a multicentre study. Eur Heart J Cardiovasc Imaging 16(4):410–416
Yingchoncharoen T et al (2013) Normal ranges of left ventricular strain: a meta-analysis. J Am Soc Echocardiogr 26(2):185–191
Xia JZ et al (2014) Left ventricular strain examination of different aged adults with 3D speckle tracking echocardiography. Echocardiography 31(3):335–339
Xu T-Y et al (2014) Three-dimensional speckle strain echocardiography is more accurate and efficient than 2D strain in the evaluation of left ventricular function. Int J Cardiol 176(2):360–366
Ringle A et al (2017) Evolution of subclinical myocardial dysfunction detected by two-dimensional and three-dimensional speckle tracking in asymptomatic type 1 diabetic patients: a longterm follow-up study. Echo Res Pract 4(4):73–81
Rozenbaum Z et al (2019) Association of body mass index and diastolic function in metabolically healthy obese with preserved ejection fraction. Int J Cardiol 277:147–152
Gandhi S et al (2018) Automation, machine learning, and artificial intelligence in echocardiography: a brave new world. Echocardiography 35(9):1402–1418
Funding
Dr. Ferit Onur Mutluer is the recipient of a Turkish Society of Cardiology Research Grant for the year 2019 and an European Association of Cardiovascular Imaging (EACVI) research grant for the year 2020.
Author information
Authors and Affiliations
Contributions
Conceptualization: JRH, AVB; Methodology: JRH, AVB; Formal analysis and investigation: FOM, DB, RVG, AVB; Writing—original draft preparation: FOM, DB, AVB; Writing—review and editing: FOM, DB, RVG, AVB, JRH; Funding acquisition: AVB, JRH; Resources: JRH, AVB; Supervision: JRH, AVB.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
All procedures were in accordance with the ethical standards of the institutional research committee and with the Helsinki declaration and its later amendments.
Informed consent
Written informed consent was obtained from the subjects prior to participation in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mutluer, F.O., Bowen, D.J., van Grootel, R.W.J. et al. Left ventricular strain values using 3D speckle-tracking echocardiography in healthy adults aged 20 to 72 years. Int J Cardiovasc Imaging 37, 1189–1201 (2021). https://doi.org/10.1007/s10554-020-02100-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10554-020-02100-3