Abstract
Purpose
Estrogen Receptor α (ERα) is a well-established therapeutic target for Estrogen Receptor (ER)-positive breast cancers. Both Selective Estrogen Receptor Degraders (SERD) and PROTAC ER degraders are synthetic compounds suppressing the ER activity through the degradation of ER. However, the differences between SERD and PROTAC ER degraders are far from clear.
Methods
The effect of PROTAC ER degrader ERD-148 and SERD fulvestrant on protein degradation was evaluated by western blot analysis. The cell proliferation was tested by WST-8 assays and the gene expressions were assessed by gene microarray and real-time RT-PCR analysis after the compound treatment.
Results
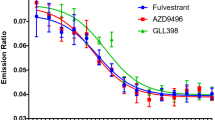
ERD-148 is a potent and selective PROTAC ERα degrader. It degrades not only unphosphorylated ERα but also the phosphorylated ERα in the cells. In contrast, the SERD fulvestrant showed much-reduced degradation potency on the phosphorylated ERα. The more complete degradation of ERα by ERD-148 translates into a greater maximum cell growth inhibition. However, ERD-148 and fulvestrant share a similar gene regulation profile except for the variation of regulation potency. Further studies indicate that ERD-148 degrades the ERα in fulvestrant-resistant cells.
Conclusion
PROTAC ER degrader has a different mechanism of action compared to SERD which may be used in treating fulvestrant-resistant cancers.











Similar content being viewed by others
Data availability
Data are available upon request.
Abbreviations
- ER:
-
Estrogen receptor
- ERD:
-
Estrogen receptor degrader
- FITC:
-
Fluorescein-5-isothiocyanate
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- Imax:
-
Maximum inhibition
- PCR:
-
Polymerase chain reaction
- PI:
-
Propidium iodide
- PROTAC:
-
Proteolysis-targeting chimeras
- PVDF:
-
Polyvinylidene difluoride
- RIPA buffer:
-
Radioimmunoprecipitation assay buffer
- SERD:
-
Selective estrogen receptor degraders
- SERM:
-
Selective estrogen receptor modulators
References
Bjornstrom L, Sjoberg M (2005) Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol 19:833–842
Fuentes N, Silveyra P (2019) Estrogen receptor signaling mechanisms. Adv Protein Chem Struct Biol 116:135–170
Lumachi F, Brunello A, Maruzzo M, Basso U, Basso SM (2013) Treatment of estrogen receptor-positive breast cancer. Curr Med Chem 20:596–604
Jia M, Dahlman-Wright K, Gustafsson JA (2015) Estrogen receptor alpha and beta in health and disease. Best Pract Res Clin Endocrinol Metab 29:557–568
Paterni I, Granchi C, Katzenellenbogen JA, Minutolo F (2014) Estrogen receptors alpha (ERalpha) and beta (ERbeta): subtype-selective ligands and clinical potential. Steroids 90:13–29
Kargbo RB (2020) Selective estrogen receptor degraders for the potential treatment of cancer. ACS Med Chem Lett 11:412–413
Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P (1995) Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270:1491–1494
Lannigan DA (2003) Estrogen receptor phosphorylation. Steroids 68:1–9
Patel HK, Bihani T (2018) Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol Ther 186:1–24
Wang L, Guillen VS, Sharma N, Flessa K, Min J, Carlson KE, Toy W, Braqi S, Katzenellenbogen BS, Katzenellenbogen JA, Chandarlapaty S, Sharma A (2018) New class of selective estrogen receptor degraders (SERDs): expanding the toolbox of PROTAC degrons. ACS Med Chem Lett 9:803–808
Hernando C, Ortega-Morillo B, Tapia M, Moragon S, Martinez MT, Eroles P, Garrido-Cano I, Adam-Artigues A, Lluch A, Bermejo B, Cejalvo JM (2021) Oral selective estrogen receptor degraders (SERDs) as a novel breast cancer therapy: present and future from a clinical perspective. Int J Mol Sci 22(15):7812
Pike AC, Brzozowski AM, Walton J, Hubbard RE, Thorsell AG, Li YL, Gustafsson JA, Carlquist M (2001) Structural insights into the mode of action of a pure antiestrogen. Structure 9:145–153
Weir HM, Bradbury RH, Lawson M, Rabow AA, Buttar D, Callis RJ, Curwen JO, de Almeida C, Ballard P, Hulse M, Donald CS, Feron LJ, Karoutchi G, MacFaul P, Moss T, Norman RA, Pearson SE, Tonge M, Davies G, Walker GE, Wilson Z, Rowlinson R, Powell S, Sadler C, Richmond G, Ladd B, Pazolli E, Mazzola AM, D’Cruz C, De Savi C (2016) AZD9496: an oral estrogen receptor inhibitor that blocks the growth of ER-positive and ESR1-mutant breast tumors in preclinical models. Cancer Res 76:3307–3318
Zhang X, Zhang Z, Xue X, Fan T, Tan C, Liu F, Tan Y, Jiang Y (2022) PROTAC degrader of estrogen receptor alpha targeting DNA-binding domain in breast cancer. ACS Pharmacol Transl Sci 5:1109–1118
Hu J, Hu B, Wang M, Xu F, Miao B, Yang CY, Wang M, Liu Z, Hayes DF, Chinnaswamy K, Delproposto J, Stuckey J, Wang S (2019) Discovery of ERD-308 as a highly potent proteolysis targeting chimera (PROTAC) degrader of estrogen receptor (ER). J Med Chem 62:1420–1442
Kargbo RB (2019) PROTAC-mediated degradation of estrogen receptor in the treatment of cancer. ACS Med Chem Lett 10:1367–1369
Wang Z, Ma Z, Shen Z (2021) Selective degradation of the estrogen receptor in the treatment of cancers. J Steroid Biochem Mol Biol 209:105848
Qin H, Zhang Y, Lou Y, Pan Z, Song F, Liu Y, Xu T, Zheng X, Hu X, Huang P (2022) Overview of PROTACs targeting the estrogen receptor: achievements for biological and drug discovery. Curr Med Chem 29:3922–3944
Jimenez DG, Sebastiano MR, Caron G, Ermondi G (2021) Are we ready to design oral PROTACs(R)? ADMET DMPK 9:243–254
Lin X, Xiang H, Luo G (2020) Targeting estrogen receptor alpha for degradation with PROTACs: a promising approach to overcome endocrine resistance. Eur J Med Chem 206:112689
Liu Z, Hu M, Yang Y, Du C, Zhou H, Liu C, Chen Y, Fan L, Ma H, Gong Y, Xie Y (2022) An overview of PROTACs: a promising drug discovery paradigm. Mol Biomed 3:46
Qi SM, Dong J, Xu ZY, Cheng XD, Zhang WD, Qin JJ (2021) PROTAC: an effective targeted protein degradation strategy for cancer therapy. Front Pharmacol 12:692574
Wang C, Zhang Y, Wu Y, Xing D (2021) Developments of CRBN-based PROTACs as potential therapeutic agents. Eur J Med Chem 225:113749
Xi JY, Zhang RY, Chen K, Yao L, Li MQ, Jiang R, Li XY, Fan L (2022) Advances and perspectives of proteolysis targeting chimeras (PROTACs) in drug discovery. Bioorg Chem 125:105848
Gonzalez TL, Hancock M, Sun S, Gersch CL, Larios JM, David W, Hu J, Hayes DF, Wang S, Rae JM (2020) Targeted degradation of activating estrogen receptor alpha ligand-binding domain mutations in human breast cancer. Breast Cancer Res Treat 180:611–622
Tan H, Zhong Y, Pan Z (2009) Autocrine regulation of cell proliferation by estrogen receptor-alpha in estrogen receptor-alpha-positive breast cancer cell lines. BMC Cancer 9:31
Hu B, Wu Z, Bai D, Liu T, Ullenbruch MR, Phan SH (2015) Mesenchymal deficiency of Notch1 attenuates bleomycin-induced pulmonary fibrosis. Am J Pathol 185:3066–3075
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264
Parrish RS, Spencer HJ 3rd (2004) Effect of normalization on significance testing for oligonucleotide microarrays. J Biopharm Stat 14:575–589
Joel PB, Traish AM, Lannigan DA (1998) Estradiol-induced phosphorylation of serine 118 in the estrogen receptor is independent of p42/p44 mitogen-activated protein kinase. J Biol Chem 273:13317–13323
Arnold SF, Vorojeikina DP, Notides AC (1995) Phosphorylation of tyrosine 537 on the human estrogen receptor is required for binding to an estrogen response element. J Biol Chem 270:30205–30212
Acknowledgements
We thank the assistance of the Department of Internal Medicine at the University of Michigan for this study.
Funding
This study is supported by the University of Michigan Regional Comprehensive Metabolomics Resource Core Pilot and feasibility Grant (U24 DK097153).
Author information
Authors and Affiliations
Contributions
BH designed and conducted the experiments, analyzed and interpreted the data, and wrote the manuscript. JH conducted the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. The use of animals and cell lines was conducted following the NIH guidelines in the USA.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, B., Hu, J. Complete elimination of estrogen receptor α by PROTAC estrogen receptor α degrader ERD-148 in breast cancer cells. Breast Cancer Res Treat 203, 383–396 (2024). https://doi.org/10.1007/s10549-023-07136-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-07136-2