Abstract
Purpose
ROR1 and ROR2 are Type 1 tyrosine kinase-like orphan receptors for Wnt5a that are associated with breast cancer progression. Experimental agents targeting ROR1 and ROR2 are in clinical trials. This study evaluated whether expression levels of ROR1 or ROR2 correlated with one another or with clinical outcomes.
Methods
We interrogated the clinical significance of high-level gene expression of ROR1 and/or ROR2 in the annotated transcriptome dataset from 989 patients with high-risk early breast cancer enrolled in one of nine completed/graduated/experimental and control arms in the neoadjuvant I-SPY2 clinical trial (NCT01042379).
Results
High ROR1 or high ROR2 was associated with breast cancer subtypes. High ROR1 was more prevalent among hormone receptor-negative and human epidermal growth factor receptor 2-negative (HR-HER2-) tumors and high ROR2 was less prevalent in this subtype. Although not associated with pathologic complete response, high ROR1 or high ROR2 each was associated with event-free survival (EFS) in distinct subtypes. High ROR1 associated with a worse EFS in HR + HER2- patients with high post-treatment residual cancer burden (RCB-II/III) (HR 1.41, 95% CI = 1.11–1.80) but not in patients with minimal post-treatment disease (RCB-0/I) (HR 1.85, 95% CI = 0.74–4.61). High ROR2 associated with an increased risk of relapse in patients with HER2 + disease and RCB-0/I (HR 3.46, 95% CI = 1.33–9.020) but not RCB-II/III (HR 1.07, 95% CI = 0.69–1.64).
Conclusion
High ROR1 or high ROR2 distinctly identified subsets of breast cancer patients with adverse outcomes. Further studies are warranted to determine if high ROR1 or high ROR2 may identify high-risk populations for studies of targeted therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
ROR1 encodes a developmentally restricted type I receptor tyrosine kinase-like orphan receptor, [1,2,3,4] which we identified was a receptor of Wnt5a. [5] ROR1 expression is prominent in embryogenesis, attenuates during fetal development, and is minimal in post-partum tissues. However, ROR1 is expressed by neoplastic cells of many cancer types making it a potential target for cancer therapy [5, 6]. High-level expression of ROR1 on breast cancer cells has been associated with epithelial–mesenchymal transition (EMT), tumor cell proliferation, and metastases [7]. In chronic lymphocytic leukemia (CLL), high-level expression of ROR1 associates with more-rapid disease progression and shorter survival. [8] As such, the expression of ROR1 may have functional significance that can influence clinical outcomes.
ROR2 encodes another developmentally restricted, type I tyrosine kinase-like orphan receptor that is structurally related to ROR1 and can serve as a receptor for Wnt5a. [9] Recent studies suggest that ROR2 signaling also may contribute to breast cancer progression and/or tissue invasiveness. [10] It is not known whether the expression of ROR2 correlates with expression of ROR1, with specific breast cancer subtypes, or with differences in clinical outcomes.
We examined the relationship between gene expression of ROR1 and/or ROR2 and outcomes in breast cancer patients enrolled in the “Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging And Molecular Analysis 2” (I-SPY2 TRIAL) study. I-SPY2 is an adaptive platform for investigating novel agents for neoadjuvant treatment of high-risk patients with poor prognosis, newly diagnosed early breast cancer. [11, 12] I-SPY2 employs clinical biomarkers to classify patients’ tumors into subtypes, allowing for randomization of patients into groups that can undergo treatment with or without novel agents proposed for neoadjuvant therapy. Pretreatment transcriptome data are collected on each tumor sample, which is annotated with biomarker subtypes into subgroups that have disparate clinical outcomes. These data can inform development of new therapeutics for patients with resistant disease. In this study, we interrogated the I-SPY2 clinical and transcriptome dataset to determine whether the gene expression levels of ROR1 or ROR2 at diagnosis, alone or together, correlate with clinical subtype, response to neoadjuvant chemotherapy, or event-free survival (EFS).
Although expression of ROR1 or ROR2 transcripts generally correlate with the expression of ROR1 or ROR2 protein [13], studies have identified alternative splice variants of each of these genes that appear unable to encode surface proteins [14], making this correlation tentative. Nonetheless, platforms for interrogating the genes expressed in breast cancers increasingly are being used to identify subtypes of this disease that have prognostic value. We hypothesize that prognostic value also may be observed in stratifying breast cancers with respect to their relative levels of ROR1 and/or ROR2 in the context of residual disease or associated clinical subtype.
Materials and methods
Patients and the I-SPY2 trial
We interrogated the clinical significance of high-level gene expression of ROR1 and/or ROR2 in 989 patients with stage II or III breast cancer and high-risk disease by clinical criteria (HR- HER2- or HER2 +) or high-risk disease according to the 70-gene signature. [15] Patients were enrolled in one of nine completed/graduated/experimental and control arms in the multi-center, multi-arm neoadjuvant I-SPY2 clinical trial (NCT01042379, IND 105,139) as depicted in Supplemental Fig. 1. Detailed descriptions of the I-SPY2 study design, eligibility, and assessments are as reported [16,17,18,19,20,21,22].
Distribution of ROR1 and ROR2 expression by HR/HER2 subtype. a, b Violin plots of log2-scaled normalized a ROR1 and b ROR2 expression by HR and HER2 status. Asterisks reflects pairwise Wilcoxon-rank sum test p values (*** p < 0.0001, **0.0001 < p < 0.001, *0.001 < p < 0.05). Color denotes receptor subtype (pink: HR + HER2-, green: HR + HER2 +, aqua: HR- HER2-, purple: HR-HER2) c Scatter plot of ROR2 vs. ROR1 expression level. Color reflects receptor subtype (pink: HR + HER2-, green: HR + HER2 +, aqua: HR- HER2-, purple: HR- HER2+). Dotted lines indicate median ROR1 and ROR2 expression values, which were used to define four patient subsets by dichotomized ROR1 and ROR2 expression (ROR1 above median: ROR1-High; ROR2 above median: ROR2-High). d Distribution of dichotomized ROR1/ROR2 expression subsets by HR and HER2 status. Color reflects ROR1/ ROR2 expression groups (red: ROR1-High/ROR2-High (HH); orange: ROR1-High/ROR2-Low (HL); light blue: ROR1-Low/ROR2-High (LH); blue: ROR1-Low/ROR2-Low (LL)
Ethics
Institutional Review Boards at all participant institutions approved the protocol. All patients provided signed informed consent to allow for research on their biospecimen samples in association with clinical outcome data.
Datasets
Platform corrected, log2-transformed, and normalized gene-level transcriptomic data generated from pretreatment tumor samples assayed on Agilent 44 K expression arrays were obtained from NCBI’s Gene Expression Omnibus (GEO) (GSE194040). We obtained patient-level scores from expression signatures reflecting estrogen receptor signaling, HER2 signaling, and proliferation from the supplemental data of the associated publication. [22]
Statistical analysis
We assessed association between ROR1 or ROR2 gene expression levels and hormone receptor (HR) and human epidermal growth factor receptor 2 (HER2) defined subtypes using a Kruskal–Wallis test with post hoc pairwise comparisons by Wilcoxon-rank sum tests with default (Holm) adjustment for multiple hypothesis testing. We used logistic regression to assess association between ROR1 or ROR2 expression levels and pathologic complete response (pCR) with significance assessment, using the likelihood ratio test comparing models with or without the biomarker term. We performed analyses with univariate and multivariate models, adjusted for subtype and treatment, conducted within-subtype analyses, with and without adjusting for treatment, as well as exploratory analyses within subtype and within arm. We used multivariate Cox proportional hazard modeling to assess association between ROR1/ROR2 expression levels and EFS with significance assessment, using the likelihood ratio test (comparing models with/without the biomarker term). These analyses were performed in the overall population adjusting for subtype and treatment and extent of residual disease (RCB-0/I vs. RCB-II/III [23]), and among RCB-0/I and RCB-II/III patients, adjusting for subtype and treatment; within-subtype analyses adjusting for treatment, for treatment and extent of residual disease, and among RCB-0/I and RCB-II/III patients. Association between ROR2 expression levels and subtype, pCR, and EFS were similarly evaluated. Pearson correlation was used to assess correlations between the expression levels of ROR1 and ROR2 with expression levels of EMT-related pathway genes, including Hippo/Yap/TAZ, WNT5A, BMl1, BCL2, and GLI1, as well as two ER-related, two HER2-related, and two proliferation-related expression signatures. In addition, we also compared expression levels of these genes and signatures between patient subsets defined by ROR1 and ROR2 expression levels (above versus below the median) using the ANOVA F test. All analyses were performed using R version 3.6.3 without adjustment for multiple hypotheses testing.
The analysis reported here is a biomarker study of the gene expression of ROR1 and ROR2 leveraging data from the I-SPY2 clinical trial. The patient population, specimen collection, assay methods, and trial design were all previously described, and the sample size could not be changed for this study. REMARK criteria were used to report the data. [24].
Results
Expression of ROR1 and ROR2 in breast cancer subtypes
We examined the expression levels of ROR1 at baseline by subtype of breast cancer, Fig. 1a. We observed a wide range of ROR1 expression levels in all subtypes. We noted HR- HER2- breast cancers expressed the highest levels of ROR1, followed by cancers with the HR-HER2 + subtype. HR + HER2- tumors expressed lower levels, which were not significantly different from that of HR + HER2 + tumors. In contrast to what we found for ROR1, the expression levels of ROR2 were lowest in HR-HER2- breast cancers and significantly lower than that found in other subtypes; the highest ROR2 expression levels were observed in the HR + HER2 + subtype, Fig. 1b. A weak-positive correlation was observed between the expression levels of ROR1 and ROR2 , Fig. 1c. When dichotomized at the median expression levels of ROR1 and ROR2 and divided into 4 subgroups, we observed an association between the subgroups defined by high-level expression of ROR1 and ROR2 in breast cancer subtypes, Fig. 1d. The choice of median cut-point for high- versus low-level expression was based upon our prior study using the median cut-point in analyzing ROR1 expression in breast cancer datasets before and after neoadjuvant treatment. [25, 26] High-level expression of ROR1 and ROR2 was noted in a large percentage of HER2 + specimens, whereas high-level ROR1 and low-level ROR2 were more common in HR- HER2- tumors. Consistent with the enrichment for HR- HER2- tumors, the subset with high ROR1 and low ROR2 has the lowest expression levels of ER- and HER2-related signatures and the highest gene expression signatures associated with proliferation, Supplemental Table S1.
Expression of ROR1 and ROR2 and likelihood of pCR
Analysis of likelihood of pCR (RCB-0) [23] by overall population and by subtype revealed a wide range of ROR1 expression levels within both pCR and non-pCR groups. Although higher expression levels of ROR1 associated with non-pCR in the HR- HER2- subtype, Fig. 2a, the higher ROR1 expression observed in non-pCR patients did not retain significance when adjusted for treatment arm. Moreover, there was no apparent association with pCR in other breast cancer subtypes, Supplemental Table S2. Exploratory analysis of pCR in HR-HER2- patients by treatment arm indicated a trend toward negative association of high ROR1 expression and pCR in 5 of the 8 treatment arms with a notably strong signal in the 32 patients treated on the MK2206 (AKT inhibitor) arm, Fig. 2b. Analysis of ROR2 expression in relationship to pCR revealed that high-level ROR2 was not associated with pCR in the overall population or in any subtype, Supplemental Table S2. Therefore, neither high-level ROR1 nor high-level ROR2 was associated with the likelihood of pCR.
Association between ROR1 and pCR within HR/HER2 subtypes. a Violin plot of ROR1 expression by pCR and HR, HER2 status. Color reflects pCR status; asterisk denotes likelihood ratio test p < 0.05. b Forest plot showing odds ratio of achieving a pCR associated with 1 standard deviation increase of ROR1 expression among HR-HER2- patients (overall, overall adjusting for treatment, and within each treatment arm)
Association of ROR1 / ROR2 and EFS
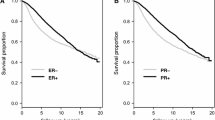
Breast cancers from patients who had high-level expression of ROR1 had a worse EFS when adjusted for subtype and treatment arm (HR 1.2, 95% CI = 1.03–1.40, LRp = 0.02), Table 1. When assessed in the context of residual disease, high-level expression of ROR1 associated with a significantly worse outcome for patients with HR + HER2- tumors who had a high post-treatment residual cancer burden (RCB-II/III) (HR = 1.41, 95% CI = 1.11–1.80, LRp = 0.01). However, we did not observe a significant association between EFS and ROR1 among patients with HR + HER2- tumors who had little or no post-treatment residual cancer burden (RCB-0/I) (HR = 1.85, 95% CI = 0.74–4.61, LRp = 0.19), which may in part be attributable to a smaller number of events within the RCB-0/I group. We did not observe an association between high-level expression of ROR1 and worse EFS among patients with HR-HER2- or HER2 + cancer subtypes. Inclusion of ROR2 in the analysis model did not change these findings. Kaplan–Meier exploratory analysis by ROR1 above or below the median level indicated that HR + HER2- patients with high-level ROR1 at baseline and high-tumor burden after treatment (RCB-II/III) had significantly worse EFS, (HR = 0.55, 95% CI = 0.33–0.9, LRp = 0.02), Fig. 3a. Kaplan–Meier plots further stratified by RCB class showed that high ROR1 in HR + HER2- patients with RCB-III had the worst outcome, Supplemental Figure S2a.
Association between ROR1 and ROR2 expression and event-free survival in the context of subtypes and extent of residual disease. Kaplan–Meier plots of a HR + HER2- patients with moderate and significant residual disease (RCB-II/III) dichotomized by median ROR1 expression (purple: below median; orange: above median) and b HER2 + patients with no or minimal residual disease (RCB-0/I) dichotomized by median ROR2 expression (purple: below median; orange: above median)
Patients with breast cancers exhibiting high-level expression of ROR2 did not have a significant difference in EFS compared to patients with tumors with low-level expression of ROR2 when adjusted for subtype and treatment arm (HR = 1.09, 95% CI = 0.94–1.27, LRp = 0.27), Table 1. However, after adjustment for subtype, treatment, and RCB class, patients with HER2 + subtype tumors and minimal residual disease after treatment (RCB-0/I) had significantly worse EFS (HR = 3.46, 95% CI = 1.33–9.02, LRp = 0.01,) Table 1, if their breast cancers expressed high levels of ROR2. Inclusion of ROR1 in this analysis model did not change these findings but provided for a numerically larger hazard ratio in the HER2 + RCB-0/I group (HR = 4.87, 95%CI = 1.57–15.09, LRp = 0.004). Kaplan–Meier analysis of EFS by ROR2 above or below the median revealed that, among patients who had little or no residual disease after therapy (RCB-0/I), those with HER2 + tumors and high-level expression of ROR2 at baseline had a significantly worse EFS than those with HER2 + tumors and low levels of ROR2 (HR = 0, 95% CI 0-Inf, LRp = 0.01), Fig. 3b. Further stratification by RCB class showed that high ROR2 HER2 + patients with RCB-0 or RCB-I had similar EFS, Supplemental Figure S2b. Analysis of EFS by ROR2 in the RCB-0 (pCR) group with only 6 events did not show a significant difference, Supplemental Fig. 2b. Further exploratory evaluation of 905 I-SPY2 patients with follow-up information regarding recurrence status and site of recurrent disease did not reveal a significant association between high-level expression of ROR2 and the occurrence of isolated CNS metastases (N = 22) or the occurrence of CNS metastases in combination with metastases at other sites (N = 18) (data not shown).
Expression of ROR1 associates with high-level expression of EMT-related genes
We performed hierarchical clustering of ROR1 with 24 EMT-related genes including the Hippo signaling pathway genes from the MSigDB database [27] along with WNT5a, BMI1, BCL2, and GLI1 prompted by associations noted in prior studies on breast cancer or CLL [25, 28,29,30]. As shown in Fig. 4, we noted a significant association between the high-level expression of ROR1 and 20 genes evaluated. Eighteen genes each had a positive correlation with ROR1: WWTR1; AMOTL1; AMOT; LATS2; YAP1; SAV1; LATS1; ROR2; GLI1; AMOTL2; NPHP4; MOB1B; WNT5A; DVL2; TJP2; STK4; MOB1A; and TJP1. Two genes each had a significant negative correlation with ROR1: BCL2 and YWHAB. The strongest correlation between ROR1 and an EMT-related gene was with WWTR1 (TAZ) (Rp = 0.54), Supplemental Table S3.
Correlation plot of ROR1 and ROR2 expression with EMT-related genes. Genes are organized by hierarchical clustering based on Pearson correlation. Color intensity of the dot reflects the magnitude of Pearson correlation coefficient (red: positive, blue: negative). Size of the dot reflects the p value, and x marks correlations with p > 0.05
Similarly, 18 EMT-related genes had a positive correlation with ROR2: GLI1; BCL2; STK4; YWHAB; AMOT; LATS1; AMOTL1; TJP2; WWC1; NPHP4; DVL2; YAP1; ROR1; WWTR1; WNT5a; TJP1; AMOTL2; and LATS2. Only 2 genes had a significant negative correlation with ROR2: MOB1A and STK3. The strongest correlation between ROR2 and an EMT-related gene was with LATS1 (Rp = 0.57), Supplemental Table S3. Consistent with the correlation analysis, comparison of expression levels of EMT genes between the four ROR1/ROR2 groups defined using ROR1/ROR2 expression, 21 of 24 EMT-related genes assessed were differentially expressed between these groups, Supplemental Table S3.
Discussion
Using the annotated I-SPY2 transcriptome data from a cohort of nearly 1000 patients with newly diagnosed high-risk early breast cancer, we found that high-level pretreatment expression of ROR1 or ROR2 had a distinct subtype-specific association with adverse risk. High-level expression of ROR1 was highest in HR- HER2- subtype and was associated with worse EFS in HR + HER2- patients with high post-treatment residual cancer burden (RCB-II/III). High-level expression of ROR2 was lowest in the HR- HER2- subtype of breast cancers and higher ROR2 expression was associated with worse EFS in HER2 + patients with minimal residual disease after therapy (RCB-0/I). High-level ROR1 or high-level ROR2 each was associated with high-level expression of genes involved in EMT. Although not correlated with pCR, high-level expression of ROR1 or ROR2 distinctly identified breast cancer patients with different tumor subtypes with adverse outcomes. This study highlights the potential prognostic value in assessing the levels of ROR1 and/or ROR2 in untreated high-risk early-stage breast cancer and justifies further studies to evaluate the biology and possible value of targeting ROR1 and ROR2 with investigational treatments.
Prior studies from our group showed an association of ROR1 signaling with stem cell features, EMT, proliferation, and metastases in preclinical models; moreover, the apparent reversal of such features by treatment with an inhibitory anti-ROR1 antibody justified correlative studies of ROR1 expression with response and clinical outcome [25]. Interrogation of tumor biopsies from 122 patients before and after neoadjuvant chemotherapy revealed the expression level of ROR1 was increased in residual breast cancer cells after surgery and was associated with enhanced expression of genes associated with EMT, proliferation, and cancer stemness. [25] Therefore, studies of the expression levels of ROR1 and ROR2 on post-treatment surgical specimens in the I-SPY2 transcriptome dataset, when it becomes available, may provide biologic insights, inform future clinical trials, and determine the optimal tissue and timing for assay.
An exploratory analysis of pCR in HR- HER2- patients by treatment arm indicated a negative trend for the association of high-level ROR1 with pCR in 5 of the 8 treatment arms with a notably strong signal in the 32 patients treated with MK2206, an AKT inhibitor. This strong signal with MK2206 is not surprising as ROR1 signaling activates the PI3K/AKT/MTOR pathway [25] and high-level expression of ROR1 may mitigate the anti-tumor activity of an AKT inhibitor in combination with chemotherapy. This observation suggests that investigations of ROR1 blockade with inhibitors of AKT signaling may be informative.
The results for ROR2 expression significantly extend the prior observations that ROR2 signaling also may contribute to breast cancer progression and/or tissue invasiveness [9, 10]. Studies have shown that ROR2 may regulate the balance of Wnt signaling and cellular heterogeneity during tumor progression. [31] To our knowledge, our study is the first to evaluate the expression levels of ROR1 and ROR2 in the same large clinical dataset and to evaluate the association of ROR2 expression with response and EFS by subtype. Our findings that elevated ROR2 expression associated with worse outcome in HER2 + patients with minimal post-treatment residual cancer burden (RCB-0/I) was based on a small number of events. As such, analyses of additional datasets are warranted to determine if high-level expression of ROR2 is associated with adverse outcomes and to determine other factors that may impact outcome in this patient subset.
Our study has several strengths and limitations. Strengths include the fact that the I-SPY2 trial platform includes robust correlative science on serial tumor biospecimens, an active control arm, and contemporary chemotherapy backbone [19]. As a result, we were able to evaluate associations of pretreatment ROR1 and/or ROR2 with chemotherapy response by pCR and clinical outcome by EFS. Of note, I-SPY2 eligibility requires that all tumors be clinically or molecularly high risk and patient performance status be excellent. The average age of enrolled patients is more than 10 years younger than that of typical breast cancer patients. [19] Therefore this study may not reflect ROR1 and ROR2 expression in the typical patient with breast cancer.
Potential limitations of our study include the analysis of pretreatment tumor specimens only, analysis of gene expression only, and use of Agilent 44 K platform which cannot distinguish between RNA isoforms of ROR1 or ROR2 that can or cannot be expressed as cell surface proteins [14]. Additionally, multiple hypothesis testing and small numbers of events in many categories limit statistical power. We analyzed the I-SPY2 transcriptome dataset of baseline pretreatment tumor specimens for expression of ROR1 and ROR2 as a transcriptome dataset for post-neoadjuvant surgical tumor tissue is not yet available. Breast cancer biology, hormone receptors, subtype frequency, and mutations can evolve over time under the pressure of systemic therapy; therefore, pretreatment tumor specimens may have different biomarker expressions than post-treatment tumor specimens. [32] However, current biomarkers with clinical utility in early breast cancer are based on assays of pretreatment tumor specimens justifying the current investigation of pretreatment specimens. Future studies of post-treatment surgical specimens, when available, and metastatic specimens are warranted to determine the optimal timing for assessment of ROR1 and ROR2 to inform clinical trials of targeted agents.
As noted, the expression or ROR1 or ROR2 may not accurately reflect the expression of ROR1 or ROR2 protein. Therefore, we examined the expression of genes that may be upregulated in breast cancer cells through activation of ROR1 or ROR2 signaling. [25, 28] Hierarchical clustering reveals significant associations of ROR1 with 20 of 24 EMT-related genes, and the strongest association is with WWTR1 (TAZ) a transcriptional coactivator in the Hippo signaling pathway. [33] Similar hierarchical clustering analysis of ROR2 expression and EMT-related genes reveals significant associations of ROR2 with 18 of 24 EMT-related genes with the strongest association with LATS1 (Rp = 0.57), hypothesized to be a tumor suppressor and the main kinase component in the Hippo signaling pathway [34]. Our analysis showed significant, but variable, correlations between WNT5a and ROR1 or WNT5a and ROR2. This observation is expected because WNT5a gene expression can be modulated by many different pathways [35] and not exclusively by ROR1- and ROR2- regulated pathways. Additional correlation analysis of ROR1/ROR2 expression groups by median cut-point high/low status with gene signatures revealed that the high ROR1 and low ROR2 group enriched for HR- HER2- tumors had the lowest expression levels of ER- and HER2-related signatures and the highest expression levels of proliferation signatures. EMT gene and signature expression that were significantly correlated with ROR1 and/or ROR2 expression were also differentially expressed between the four ROR1/ROR2 defined subsets.
Cancer cells may express ROR1 or ROR2 at levels not observed in normal post-partem tissues and, therefore, the protein antigens encoded by these genes could serve as potential targets for therapy. [25] Our group has developed a humanized monoclonal antibody, cirmtuzumab (now designated as zilovertamab), to ROR1. [36] A Phase 1 study in CLL showed that zilovertamab therapy reversed ROR1 signaling and stemness signatures with minimum apparent toxicity. [37] As a result, zilovertamab is currently under study in CLL and mantle cell lymphoma (NCT03088878) and in advanced breast cancer (NCT02776917) with no additional safety concerns reported to date. [38, 39] Our group has also developed a ROR1 antibody conjugated to MMAE that has been found to be effective in a Richter’s syndrome mouse model [40] and this compound, VLS-101, now zilovertamab vedotin, is under study in hematologic malignancies (NCT03833180) and in solid tumors (NCT04504916). Zilovertamab vedotin was found to have no unexpected toxicities in heavily pretreated patients with lymphoid cancers and to have clinical activity [41]. Other ROR1 [42] and ROR2 targeted therapies are in clinical trials (NCT03504488, NCT03393936).
In summary, we have shown in a cohort of almost 1000 high-risk early-stage breast cancer patients treated on the I-SPY2 platform that pretreatment expression of ROR1 was higher in HR- HER2- subtype, did not correlate with pCR, and was associated with worse EFS in HR + HER2- patients with high post-treatment residual cancer burden (RCB-II/III). We found that expression of ROR2 was lowest in HR- HER2- breast cancer, highest in the HR + HER2 + subtype, and did not correlate with pCR. High ROR2 identified a subset of HER2 + patients who had an excellent response to neoadjuvant treatment (RCB-0/I) but had a higher risk of relapse. Agents targeting ROR1 and ROR2 are in clinical trials and may provide new investigational opportunities. Importantly, these results warrant further studies to determine the value of using high-level expression of ROR1 and ROR2 as markers for poor outcome that may inform clinical trials of targeted therapies.
Data availability
Platform corrected, log2-transformed, and normalized gene-level transcriptomic data generated from pretreatment tumor samples assayed on Agilent 44 K expression arrays were obtained from NCBI’s Gene Expression Omnibus (GEO) (GSE194040). As well, patient-level scores from expression signatures reflecting estrogen receptor signaling, HER2 signaling, and proliferation were obtained from the supplemental data of the associated publication. [22]
References
Masiakowski P, Carroll RD (1992) A novel family of cell surface receptors with tyrosine kinase-like domain. J Biol Chem 267(36):26181–26190
Wilson C, Goberdhan DC, Steller H (1993) Dror, a potential neurotrophic receptor gene, encodes a Drosophila homolog of the vertebrate Ror family of Trk-related receptor tyrosine kinases. Proc Natl Acad Sci U S A 90(15):7109–7113. https://doi.org/10.1073/pnas.90.15.7109
Forrester WC, Dell M, Perens E, Garriga G. (1999) A C. elegans Ror receptor tyrosine kinase regulates cell motility and asymmetric cell division. Nature.400(6747):881–5. https://doi.org/10.1038/23722.
Rodriguez-Niedenführ M, Pröls F, Christ B. (2004) Expression and regulation of ROR-1 during early avian limb development. Anat Embryol (Berl).207(6):495–502. https://doi.org/10.1007/s00429-004-0381-6.
Fukuda T, Chen L, Endo T, Tang L, Lu D, Castro JE et al (2008) Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc Natl Acad Sci U S A 105(8):3047–3052. https://doi.org/10.1073/pnas.0712148105
Zhang S, Chen L, Cui B, Chuang HY, Yu J, Wang-Rodriguez J, et al. (2012) ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS One.7(3):e31127. doi: https://doi.org/10.1371/journal.pone.0031127.
Cui B, Zhang S, Chen L, Yu J, Widhopf GF, Fecteau JF et al (2013) Targeting ROR1 inhibits epithelial-mesenchymal transition and metastasis. Cancer Res 73(12):3649–3660. https://doi.org/10.1158/0008-5472.CAN-12-3832
Cui B, Ghia EM, Chen L, Rassenti LZ, DeBoever C, Widhopf GF et al (2016) High-level ROR1 associates with accelerated disease progression in chronic lymphocytic leukemia. Blood 128(25):2931–2940. https://doi.org/10.1182/blood-2016-04-712562
Henry C, Quadir A, Hawkins NJ, Jary E, Llamosas E, Kumar D et al (2015) Expression of the novel Wnt receptor ROR2 is increased in breast cancer and may regulate both β-catenin dependent and independent Wnt signalling. J Cancer Res Clin Oncol 141(2):243–254. https://doi.org/10.1007/s00432-014-1824-y
Bayerlova M, Menck K, Klemm F, Wolff A, Pukrop T, Binder C et al (2017) Ror2 Signaling and Its Relevance in Breast Cancer Progression. Front Oncol 7:135. https://doi.org/10.3389/fonc.2017.00135
Barker AD, Sigman CC, Kelloff GJ, Hylton NM, Berry DA, Esserman LJ (2009) I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther 86(1):97–100. https://doi.org/10.1038/clpt.2009.68
Wang H, Yee D (2019) I-SPY 2: a Neoadjuvant Adaptive Clinical Trial Designed to Improve Outcomes in High-Risk Breast Cancer. Curr Breast Cancer Rep 11(4):303–310. https://doi.org/10.1007/s12609-019-00334-2
Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER, Kalocsay M et al (2020) Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 180(2):387-402.e16. https://doi.org/10.1016/j.cell.2019.12.023
John M, Ford CE. (2022) Pan-Tissue and -Cancer Analysis of ROR1 and ROR2 Transcript Variants Identify Novel Functional Significance for an Alternative Splice Variant of ROR1. Biomedicines.10(10). https://doi.org/10.3390/biomedicines10102559.
Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, et al (2016) 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med 375(8):717–729. https://doi.org/10.1056/NEJMoa1602253
Park JW, Liu MC, Yee D, Yau C, van ’t Veer LJ, Symmans WF, et al (2016) Adaptive Randomization of Neratinib in Early Breast Cancer. N Engl J Med 375(1):11–22. https://doi.org/10.1056/NEJMoa1513750
Rugo HS, Olopade OI, DeMichele A, Yau C, van ’t Veer LJ, Buxton MB, et al (2016) Adaptive Randomization of Veliparib-Carboplatin Treatment in Breast Cancer. N Engl J Med 375(1):23–34. https://doi.org/10.1056/NEJMoa1513749
Chien AJ, Tripathy D, Albain KS, Symmans WF, Rugo HS, Melisko ME et al (2020) MK-2206 and Standard Neoadjuvant Chemotherapy Improves Response in Patients With Human Epidermal Growth Factor Receptor 2-Positive and/or Hormone Receptor-Negative Breast Cancers in the I-SPY 2 Trial. J Clin Oncol 38(10):1059–1069. https://doi.org/10.1200/JCO.19.01027
Yee D, DeMichele AM, Yau C, Isaacs C, Symmans WF, Albain KS et al (2020) Association of Event-Free and Distant Recurrence-Free Survival With Individual-Level Pathologic Complete Response in Neoadjuvant Treatment of Stages 2 and 3 Breast Cancer: Three-Year Follow-up Analysis for the I-SPY2 Adaptively Randomized Clinical Trial. JAMA Oncol 6(9):1355–1362. https://doi.org/10.1001/jamaoncol.2020.2535
Nanda R, Liu MC, Yau C, Shatsky R, Pusztai L, Wallace A et al (2020) Effect of Pembrolizumab Plus Neoadjuvant Chemotherapy on Pathologic Complete Response in Women With Early-Stage Breast Cancer: An Analysis of the Ongoing Phase 2 Adaptively Randomized I-SPY2 Trial. JAMA Oncol 6(5):676–684. https://doi.org/10.1001/jamaoncol.2019.6650
Pusztai L, Yau C, Wolf DM, Han HS, Du L, Wallace AM et al (2021) Durvalumab with olaparib and paclitaxel for high-risk HER2-negative stage II/III breast cancer: Results from the adaptively randomized I-SPY2 trial. Cancer Cell 39(7):989–98.e5. https://doi.org/10.1016/j.ccell.2021.05.009
Wolf DM, Yau C, Wulfkuhle J, Brown-Swigart L, Gallagher IR, Lee PRE, et al. (2022) Redefining breast cancer subtypes to guide treatment prioritization and maximize response: Predictive biomarkers across 10 cancer therapies. Cancer Cell.40(June 13, 2022):1–15. https://doi.org/10.1016/j.ccell.2022.05.005
Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V et al (2007) Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 25(28):4414–4422. https://doi.org/10.1200/JCO.2007.10.6823
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM et al (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 100(2):229–235. https://doi.org/10.1007/s10549-006-9242-8
Zhang S, Zhang H, Ghia EM, Huang J, Wu L, Zhang J et al (2019) Inhibition of chemotherapy resistant breast cancer stem cells by a ROR1 specific antibody. Proc Natl Acad Sci U S A 116(4):1370–1377. https://doi.org/10.1073/pnas.1816262116
Chen Y, Chen L, Yu J, Ghia EM, Choi MY, Zhang L et al (2019) Cirmtuzumab blocks Wnt5a/ROR1 stimulation of NF-κB to repress autocrine STAT3 activation in chronic lymphocytic leukemia. Blood 134(13):1084–1094. https://doi.org/10.1182/blood.2019001366
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. (2011) Molecular signatures database (MSigDB) 3.0. Bioinformatics.27(12):1739–40. https://doi.org/10.1093/bioinformatics/btr260
Hasan MK, Widhopf GF, Zhang S, Lam SM, Shen Z, Briggs SP et al (2019) Wnt5a induces ROR1 to recruit cortactin to promote breast-cancer migration and metastasis. NPJ Breast Cancer 5:35. https://doi.org/10.1038/s41523-019-0131-9
Rassenti LZ, Balatti V, Ghia EM, Palamarchuk A, Tomasello L, Fadda P et al (2017) dysregulation to identify therapeutic target combinations for chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 114(40):10731–10736. https://doi.org/10.1073/pnas.1708264114
Ghia EM, Rassenti LZ, Neuberg DS, Blanco A, Yousif F, Smith EN et al (2019) Activation of hedgehog signaling associates with early disease progression in chronic lymphocytic leukemia. Blood 133(25):2651–2663. https://doi.org/10.1182/blood-2018-09-873695
Roarty K, Pfefferle AD, Creighton CJ, Perou CM, Rosen JM (2017) Ror2-mediated alternative Wnt signaling regulates cell fate and adhesion during mammary tumor progression. Oncogene 36(43):5958–5968. https://doi.org/10.1038/onc.2017.206
Cejalvo JM, Martinez de Duenas E, Galvan P, Garcia-Recio S, Burgues Gasion O, Pare L et al (2017) Intrinsic Subtypes and Gene Expression Profiles in Primary and Metastatic Breast Cancer. Cancer Res 77(9):2213–2221. https://doi.org/10.1158/0008-5472.CAN-16-2717
Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M et al (2000) TAZ: a novel transcriptional co-activator regulated by interactions with 14–3-3 and PDZ domain proteins. EMBO J 19(24):6778–6791. https://doi.org/10.1093/emboj/19.24.6778
Shen Z, Pan Y, Chen P, Jiang B, Fang X, Jiang Y (2021) LATS1 exerts tumor suppressor functions via targeting Gli1 in colorectal cancer. J Cancer 12(24):7311–7319. https://doi.org/10.7150/jca.62211
Chen YC, Gonzalez ME, Burman B, Zhao X, Anwar T, Tran M, et al. (2019) Mesenchymal Stem/Stromal Cell Engulfment Reveals Metastatic Advantage in Breast Cancer. Cell Rep.27(13):3916–26 e5. https://doi.org/10.1016/j.celrep.2019.05.084.
Choi MY, Widhopf GF, Wu CC, Cui B, Lao F, Sadarangani A et al (2015) Pre-clinical Specificity and Safety of UC-961, a First-In-Class Monoclonal Antibody Targeting ROR1. Clin Lymphoma Myeloma Leuk 15(Suppl):S167–S169. https://doi.org/10.1016/j.clml.2015.02.010
Choi MY, Widhopf GF, Ghia EM, Kidwell RL, Hasan MK, Yu J et al (2018) Phase I Trial: Cirmtuzumab Inhibits ROR1 Signaling and Stemness Signatures in Patients with Chronic Lymphocytic Leukemia. Cell Stem Cell 22(6):951–9.e3. https://doi.org/10.1016/j.stem.2018.05.018
Shatsky RA, Schwab RB, Helsten TL, Pittman EI, Chen R, Breitmeyer JB, et al. Phase 1b trial of cirmtuzumab and paclitaxel for locally advanced, unresectable and metastatic breast cancer. Proceedings of the 2019 San Antonio Breast Cancer Symposium, Cancer Res. San Antonio, TX: AACR; 2019. p. Abstract nr P3_10_8
Shatsky R, Messer K, Helsten TL, Schwab RB, Pittman EI, Chen R, et al. (2021) Phase 1b trial of cirmtuzumab and paclitaxel for locally advanced, unresectable and metastatic breast cancer. AACR Annual Virtual Meeting
Vaisitti T, Arruga F, Vitale N, Lee TT, Ko M, Chadburn A et al (2021) ROR1 targeting with the antibody drug-conjugate VLS-101 is effective in Richter syndrome patient-derived xenograft mouse models. Blood. https://doi.org/10.1182/blood.2020008404
Wang MLB, Jacqueline C, Furman RR, Mei M, Barr PM, Choi MY, de Vos S et al (2021) Zilovertamab Vedotin Targeting of ROR1 as Therapy for Lymphoid Cancers. NEJM Evidence. https://doi.org/10.1056/EVIDoa2100001
Zhao Y, Zhang D, Guo Y, Lu B, Zhao ZJ, Xu X, et al. (2021) Tyrosine Kinase ROR1 as a Target for Anti-Cancer Therapies. Front Oncol.11:680834. https://doi.org/10.3389/fonc.2021.680834
Funding
The authors acknowledge the generous support provided by the Quantum Leap Healthcare Collaborative, 2013 to present, Laura Esserman, Foundation for the National Institutes of Health, 2010 to 2012, Laura Esserman, Center for Biomedical Informatics and Information Technology, the National Cancer Institute, 28X5197, Laura Esserman, Safeway Foundation, William K. Bowles Foundation, Give Breast Cancer the Boot, Quintiles Transnational Corporation, Johnson and Johnson, Genentech, Amgen, San Francisco Foundation, Eli Lilly and Company, Pfizer, Eisai Incorporated, Side Out Foundation, Harlan Family, Avon Foundation for Women, Alexandria Real Estate Equities, Private individuals, and Family Foundations.
Author information
Authors and Affiliations
Consortia
Contributions
BAP, RAS, RBS, EMG, CY, and TJK contributed to the study conception, design, and data analysis. Material preparation and data collection were performed by AMW, I-SPY 2 Consortium, DMW, GLH, LB-S, LJE, and LJvV. The first draft of the manuscript was written by BAP and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interest
B.A.P. has received clinical research support awarded to her institution for unrelated work from Pfizer, Novartis, Glaxo Smith Kline, Genentech/Roche, and Oncternal Therapeutics Inc., received consulting fees from Dare Biosciences, had stock in Merck, and served on the San Diego Susan G. Komen Board of Directors. R.A.S. has received clinical research support awarded to her institution for unrelated work from Oncosec, Oncternal Therapeutics Inc., Phoenix Molecular Designs, Genentech/Roche, Cytomx, and OBI Pharmaceuticals; received consulting fees from the Dedham Group, Sorteria Precision Medicine Foundation, Inc., Oncosec, and Gilead; and received honoraria from San Antonio Breast Cancer Symposium, Johnson and Johnson, Total Health Conferencing, and Integrity Continuing Education. R.B.S. has received salary grant support to his institution from the sponsor of the I-SPY2 trial, Quantum Leap Healthcare Collaborative, has a pending patent Compositions and Methods for Detecting Cancer, Serial No. 13/007,23, has participated in a Cardinal Health Advisory Board, and has equity in Biosplice Therapeutics. G.L.H. and her spouse have stock in Moderna, Exact Sciences, Gilead, and Nanostring. L.B.-S. has received salary support from Quantum Leap Healthcare Collaborative for I-SPY2 operations and from the National Institutes of Health/National Cancer Institute (R01 CA255442 and P01 CA210961). L.J.E. has been an unpaid member of the Board of Directors of Quantum Leap Healthcare Collaborative (QLHC), received grant support from QLHC and from National Cancer Institute (P01) for the I-SPY2 Trial, served on an Advisory Panel for Blue Cross for which she was paid for travel and received an honorarium for her time, and receives grant support for a breast DCIS vaccine trial funded by Merck through the University of California San Francisco. L.J.v.V. has been a part-time employee and stockholder in Agendia N.V. C.Y. has received salary support to the institution from Quantum Leap Healthcare Collaborative and the National Cancer Institute. T.J.K. has received research funding for zilovertamab (previously cirmtuzumab) that was developed by T.J.K. in the T.J.K. laboratory and licensed by the University of California to Oncternal Therapeutics, Inc.; has stock options from Oncternal Therapeutics, Inc.; and has received travel funds and/or honoraria from Pharmacyclics/ AbbVie, Genentech/Roche, Janssen, Gilead, European Research Initiative on CLL (ERIC), Dava Oncology, iwNHL, NCCN CLL/ SLL Hairy Cell Leukemia Panel Meeting, Society of Hematologic Oncology. A.M.W., D.M.W., and E.M.G. have no conflicts to declare.
Ethics Approval
Approval was obtained from Institutional Review Boards at all participant institutions covering the clinics where patients were enrolled in I-SPY2 protocol as previously reported. University of California, San Francisco, Helen Diller Family Comprehensive Cancer Center, Box 1710, San Francisco, CA 94143; University of California San Diego, 3855 Health Sciences Dr. M/C 0698, La Jolla, CA 92093; University of Alabama Birmingham (UAB), Birmingham Comprehensive Cancer Center, 1802 Sixth Avenue South 2510, North Pavilion, Birmingham, AL 35294–3300; University of Minnesota (UMinn), Masonic Cancer Center, 420 Delaware St., SE, MMC 480, Minneapolis, MN 55455; Swedish Cancer Institute (Swedish), 1221 Madison Street, Seattle, WA 98104; Loyola University Medical Center, Cardinal Bernardin Cancer Center, 2160 South First Ave, Room 109, Maywood, IL 60153; Mayo Clinic Rochester (Mayo MN), 200 First St, SW, Rochester, MN 55905; University of Pennsylvania (UPenn), MSCE 3 Perelman Center 3400 Civic Center Blvd, Philadelphia, PA 19104; University of Texas, MD Anderson (MDACC), Breast Medical Oncology Dept. – Unit 1354, 1515 Holcombe Blvd., Houston, TX 77030; Georgetown University (Gtown), Lombardi Cancer Center, 3800 Reservoir Rd, NW 2nd Level Podium B, Washington, DC 20007; University of Chicago (UChi), 5841 S. Maryland Avenue, MC 2115, Chicago, IL 60437; University of Colorado (UCD), University of Colorado Cancer Center, 1665 Aurora Ct., Rm. 3200, MS F700, Aurora, CO 80045; Oregon Health and Science University, Oregon Health and Science University, 3181 SW Sam Jackson Park Rd, Portland, OR 97239; University of Texas, Southwestern (UTSW), University of Texas, Southwestern Medical Center, 5323 Harry Hines Blvd, Bldg. E6.222D, Dallas, TX 75390–9155; Moffitt Cancer Center, H. Lee Moffitt Cancer Center and Research Institute, 2902 USF Magnolia Drive, Tampa, FL 33612; University of Southern California (USC), University of Southern California, Norris Comprehensive Cancer Center, 1450 Biggy Street; Norris Research Tower (NRT) 3505, Norris Comprehensive Cancer Center, Los Angeles CA 90033; University of Arizona (UAZ), Arizona Cancer Center at UMC-North, The University of Arizona Medical Center Surgical Oncology, PO Box 245131, Tucson, AZ 85724–5131; Arizona Cancer Center at UMC, North 3838 N. Campbell Ave, Tucson, AZ 85719; University of Washington (UWash), University of Washington, 825 Eastlake Ave, East Seattle, WA 98109–1023; Inova Health System (Inova), Inova Fairfax Hospital Cancer Center, FACS 3300 Gallows Rd, Fairfax, VA 22042; Mayo Clinic Scottsdale (Mayo AZ), Mayo Clinic–Scottsdale, 13400 E. Shea Blvd, Scottsdale, AZ 85259; University of Kansas (KUMC), University of Kansas Medical Center, 2330 Shawnee Mission Pkwy, Ste 210, Westwood, KS 66205; and Emory University (Emory), Emory University Winship Cancer Institute, 1365-C Clifton Road, NE, Atlanta, GA 30322.
Consent to participate
All patients provided signed informed consent to allow for research on their biospecimen samples in association with clinical outcome data.
Consent to publish
No individual patient data or individual patient images are included in this manuscript.
Electronic figure submission
Electronic figures were generated from R version 3.6.3, assembled and formatted using Adobe Illustrator, and embedded in the text body of the manuscript in Word as requested.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Parker, B.A., Shatsky, R.A., Schwab, R.B. et al. Association of baseline ROR1 and ROR2 gene expression with clinical outcomes in the I-SPY2 neoadjuvant breast cancer trial. Breast Cancer Res Treat 199, 281–291 (2023). https://doi.org/10.1007/s10549-023-06914-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-06914-2