Abstract
Purpose
This proof-of-concept study investigates gene expression in core needle biopsies (CNB) to predict whether individuals diagnosed with ductal carcinoma in situ (DCIS) on CNB were affected by invasion at the time of diagnosis.
Methods
Using a QuantiGene Plex 2.0 assay, 14 gene expression profiling was performed in 303 breast tissue samples. Preoperative diagnostic performance of a gene was measured by area under receiver-operating characteristic curve (AUC) with 95% confidence interval (CI). The gene mRNA positivity cutoff was computed using Gaussian mixture model (GMM); protein expression was measured by immunohistochemistry; DNA methylation was evaluated by targeted bisulfite sequencing.
Results
mRNA from 69% (34/49) mammoplasties, 72% (75/104) CNB DCIS, and 89% (133/150) invasive breast cancers (IBC) were analyzed. Based on pre-and post-surgery DCIS chart reviews, 21 cases were categorized as DCIS synchronous with invasion and 54 DCIS were pure DCIS without pathologic evidence of invasive disease. The ectopic expression of neuronal cadherin CDH2 was probable in 0% mammoplasties, 6% pure DCIS, 29% synchronous DCIS, and 26% IBC. The CDH2 mRNA positivity in preoperative biopsies showing pure DCIS was predictive of a final diagnosis of invasion (AUC = 0.67; 95% CI 0.53–0.80; P = 0.029). Site-specific methylation of the CDH2 promoter (AUC = 0.76; 95% CI 0.54–0.97; P = 0.04) and measurements of N-cadherin, a pro-invasive cell–cell adhesion receptor encoded by CDH2 (AUC = 0.8; 95% CI 0.66–0.99; P < 0.005) had a discriminating power allowing for discernment of CDH2-positive biopsy.
Conclusions
Evidence of CDH2/N-cadherin expression, predictive of invasion synchronous with DCIS, may help to clarify a diagnosis and direct the course of therapy earlier in a patient’s care.




Similar content being viewed by others
Data Availability
All data needed to evaluate the conclusions in the paper are present in the paper or the supplemental information. COMPAi is available as a standalone application to qualified users.
Abbreviations
- DCIS:
-
Ductal carcinoma in situ
- CNB:
-
Core needle biopsy
- IBC:
-
Invasive breast carcinoma
- AUC:
-
Area under receiver-operating characteristic curve
- CI:
-
Confidence interval
- GMM:
-
Gaussian mixture model
- FFPE:
-
Formalin-fixed paraffin-embedded
- QG2:
-
QuantiGene Plex 2.0 Assay
- ER:
-
Estrogen receptor alpha
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor 2
- IGF-IR:
-
Insulin-like growth factor I receptor
- PPIB:
-
Peptidylprolyl isomerase B
- GUSB:
-
Glucuronidase beta
- gDNA:
-
Genomic DNA
- FDA:
-
Food and Drug Administration
- COMPAi:
-
Computer-assisted image analysis software
- ROC:
-
Receiver-operating characteristic curve
- CpG:
-
Cytosine-phosphate-guanine dinucleotides
References
Allred DC (2010) Ductal carcinoma in situ: terminology, classification, and natural history. J Natl Cancer Inst Monogr 2010(41):134–138. https://doi.org/10.1093/jncimonographs/lgq035
Gorringe KL, Fox SB (2017) Ductal carcinoma in situ biology, biomarkers, and diagnosis. Front Oncol 7:248. https://doi.org/10.3389/fonc.2017.00248
Narod SA, Sopik V (2018) Is invasion a necessary step for metastases in breast cancer? Breast Cancer Res Treat 169(1):9–23. https://doi.org/10.1007/s10549-017-4644-3
O'Sullivan MJ, Morrow M (2007) Ductal carcinoma in situ–current management. Surg Clin North Am 87(2):333–351. https://doi.org/10.1016/j.suc.2007.01.006
Benson JR, Jatoi I, Toi M (2016) Treatment of low-risk ductal carcinoma in situ: is nothing better than something? Lancet Oncol 17(10):e442–e451. https://doi.org/10.1016/S1470-2045(16)30367-9
Toss M, Miligy I, Thompson AM, Khout H, Green AR, Ellis IO, Rakha EA (2017) Current trials to reduce surgical intervention in ductal carcinoma in situ of the breast: Critical review. Breast 35:151–156. https://doi.org/10.1016/j.breast.2017.07.012
Harowicz MR, Saha A, Grimm LJ, Marcom PK, Marks JR, Hwang ES, Mazurowski MA (2017) Can algorithmically assessed MRI features predict which patients with a preoperative diagnosis of ductal carcinoma in situ are upstaged to invasive breast cancer? J Magn Reson Imaging 46(5):1332–1340. https://doi.org/10.1002/jmri.25655
Shi B, Grimm LJ, Mazurowski MA, Baker JA, Marks JR, King LM, Maley CC, Hwang ES, Lo JY (2018) Prediction of occult invasive disease in ductal carcinoma in situ using deep learning features. J Am Coll Radiol 15(3):527–534. https://doi.org/10.1016/j.jacr.2017.11.036
Brennan ME, Turner RM, Ciatto S, Marinovich ML, French JR, Macaskill P, Houssami N (2011) Ductal carcinoma in situ at core-needle biopsy: meta-analysis of underestimation and predictors of invasive breast cancer. Radiology 260(1):119–128. https://doi.org/10.1148/radiol.11102368
Kim J, Han W, Lee JW, You JM, Shin HC, Ahn SK, Moon HG, Cho N, Moon WK, Park IA, Noh DY (2012) Factors associated with upstaging from ductal carcinoma in situ following core needle biopsy to invasive cancer in subsequent surgical excision. Breast 21(5):641–645. https://doi.org/10.1016/j.breast.2012.06.012
Yen TW, Hunt KK, Ross MI, Mirza NQ, Babiera GV, Meric-Bernstam F, Singletary SE, Symmans WF, Giordano SH, Feig BW, Ames FC, Kuerer HM (2005) Predictors of invasive breast cancer in patients with an initial diagnosis of ductal carcinoma in situ: a guide to selective use of sentinel lymph node biopsy in management of ductal carcinoma in situ. J Am Coll Surg 200(4):516–526. https://doi.org/10.1016/j.jamcollsurg.2004.11.012
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of NCIEWGoCD (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 100(2):229–235. https://doi.org/10.1007/s10549-006-9242-8
Derycke LD, Bracke ME (2004) N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int J Dev Biol 48(5–6):463–476. https://doi.org/10.1387/ijdb.041793ld
Jeschke U, Mylonas I, Kuhn C, Shabani N, Kunert-Keil C, Schindlbeck C, Gerber B, Friese K (2007) Expression of E-cadherin in human ductal breast cancer carcinoma in situ, invasive carcinomas, their lymph node metastases, their distant metastases, carcinomas with recurrence and in recurrence. Anticancer Res 27(4A):1969–1974
Roses RE, Paulson EC, Sharma A, Schueller JE, Nisenbaum H, Weinstein S, Fox KR, Zhang PJ, Czerniecki BJ (2009) HER-2/neu overexpression as a predictor for the transition from in situ to invasive breast cancer. Cancer Epidemiol Biomarkers Prev 18(5):1386–1389. https://doi.org/10.1158/1055-9965.EPI-08-1101
Sanchez AM, Flamini MI, Baldacci C, Goglia L, Genazzani AR, Simoncini T (2010) Estrogen receptor-alpha promotes breast cancer cell motility and invasion via focal adhesion kinase and N-WASP. Mol Endocrinol 24(11):2114–2125. https://doi.org/10.1210/me.2010-0252
McSherry EA, Brennan K, Hudson L, Hill AD, Hopkins AM (2011) Breast cancer cell migration is regulated through junctional adhesion molecule-A-mediated activation of Rap1 GTPase. Breast Cancer Res 13(2):R31. https://doi.org/10.1186/bcr2853
Citterio C, Menacho-Marquez M, Garcia-Escudero R, Larive RM, Barreiro O, Sanchez-Madrid F, Paramio JM, Bustelo XR (2012) The rho exchange factors vav2 and vav3 control a lung metastasis-specific transcriptional program in breast cancer cells. Sci Signal 5(244):ra71. https://doi.org/10.1126/scisignal.2002962
Aguiar FN, Mendes HN, Cirqueira CS, Bacchi CE, Carvalho FM (2013) Basal cytokeratin as a potential marker of low risk of invasion in ductal carcinoma in situ. Clinics (Sao Paulo) 68(5):638–643. https://doi.org/10.6061/clinics/2013(05)010
Iyer SV, Dange PP, Alam H, Sawant SS, Ingle AD, Borges AM, Shirsat NV, Dalal SN, Vaidya MM (2013) Understanding the role of keratins 8 and 18 in neoplastic potential of breast cancer derived cell lines. PLoS ONE 8(1):e53532. https://doi.org/10.1371/journal.pone.0053532
Grassilli S, Brugnoli F, Lattanzio R, Rossi C, Perracchio L, Mottolese M, Marchisio M, Palomba M, Nika E, Natali PG, Piantelli M, Capitani S, Bertagnolo V (2014) High nuclear level of Vav1 is a positive prognostic factor in early invasive breast tumors: a role in modulating genes related to the efficiency of metastatic process. Oncotarget 5(12):4320–4336. https://doi.org/10.18632/oncotarget.2011
Guvakova MA, Lee WS, Furstenau DK, Prabakaran I, Li DC, Hung R, Kushnir N (2014) The small GTPase Rap1 promotes cell movement rather than stabilizes adhesion in epithelial cells responding to insulin-like growth factor I. Biochem J 463(2):257–270. https://doi.org/10.1042/BJ20131638
Jiang Y, Prabakaran I, Wan F, Mitra N, Furstenau DK, Hung RK, Cao S, Zhang PJ, Fraker DL, Guvakova MA (2014) Vav2 protein overexpression marks and may predict the aggressive subtype of ductal carcinoma in situ. Biomark Res 2:22. https://doi.org/10.1186/2050-7771-2-22
Afratis NA, Bouris P, Skandalis SS, Multhaupt HA, Couchman JR, Theocharis AD, Karamanos NK (2017) IGF-IR cooperates with ERalpha to inhibit breast cancer cell aggressiveness by regulating the expression and localisation of ECM molecules. Sci Rep 7:40138. https://doi.org/10.1038/srep40138
Carroll JS, Hickey TE, Tarulli GA, Williams M, Tilley WD (2017) Deciphering the divergent roles of progestogens in breast cancer. Nat Rev Cancer 17(1):54–64. https://doi.org/10.1038/nrc.2016.116
Prabakaran I, Wu Z, Lee C, Tong B, Steeman S, Koo G, Zhang PJ, Guvakova MA (2019) Gaussian Mixture Models for Probabilistic Classification of Breast Cancer. Cancer Res 79(13):3492–3502. https://doi.org/10.1158/0008-5472.CAN-19-0573
Furstenau DK, Mitra N, Wan F, Lewis R, Feldman MD, Fraker DL, Guvakova MA (2011) Ras-related protein 1 and the insulin-like growth factor type I receptor are associated with risk of progression in patients diagnosed with carcinoma in situ. Breast Cancer Res Treat 129(2):361–372. https://doi.org/10.1007/s10549-010-1227-y
Knudsen BS, Allen AN, McLerran DF, Vessella RL, Karademos J, Davies JE, Maqsodi B, McMaster GK, Kristal AR (2008) Evaluation of the branched-chain DNA assay for measurement of RNA in formalin-fixed tissues. J Mol Diagn 10(2):169–176. https://doi.org/10.2353/jmoldx.2008.070127
Groen EJ, Elshof LE, Visser LL, Rutgers EJT, Winter-Warnars HAO, Lips EH, Wesseling J (2017) Finding the balance between over- and under-treatment of ductal carcinoma in situ (DCIS). Breast 31:274–283. https://doi.org/10.1016/j.breast.2016.09.001
Toyooka KO, Toyooka S, Virmani AK, Sathyanarayana UG, Euhus DM, Gilcrease M, Minna JD, Gazdar AF (2001) Loss of expression and aberrant methylation of the CDH13 (H-cadherin) gene in breast and lung carcinomas. Cancer Res 61(11):4556–4560
Paredes J, Albergaria A, Oliveira JT, Jeronimo C, Milanezi F, Schmitt FC (2005) P-cadherin overexpression is an indicator of clinical outcome in invasive breast carcinomas and is associated with CDH3 promoter hypomethylation. Clin Cancer Res 11(16):5869–5877. https://doi.org/10.1158/1078-0432.CCR-05-0059
Caldeira JR, Prando EC, Quevedo FC, Neto FA, Rainho CA, Rogatto SR (2006) CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC Cancer 6:48. https://doi.org/10.1186/1471-2407-6-48
Hoque MO, Prencipe M, Poeta ML, Barbano R, Valori VM, Copetti M, Gallo AP, Brait M, Maiello E, Apicella A, Rossiello R, Zito F, Stefania T, Paradiso A, Carella M, Dallapiccola B, Murgo R, Carosi I, Bisceglia M, Fazio VM, Sidransky D, Parrella P (2009) Changes in CpG islands promoter methylation patterns during ductal breast carcinoma progression. Cancer Epidemiol Biomarkers Prev 18(10):2694–2700. https://doi.org/10.1158/1055-9965.EPI-08-0821
Li B, Paradies NE, Brackenbury RW (1997) Isolation and characterization of the promoter region of the chicken N-cadherin gene. Gene 191(1):7–13
Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ (1999) N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol 147(3):631–644
Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA (2000) Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol 148(4):779–790
Cavallaro U, Schaffhauser B, Christofori G (2002) Cadherins and the tumour progression: is it all in a switch? Cancer Lett 176(2):123–128
Pilewskie M, Stempel M, Rosenfeld H, Eaton A, Van Zee KJ, Morrow M (2016) Do LORIS trial eligibility criteria identify a ductal carcinoma in situ patient population at low risk of upgrade to invasive carcinoma? Ann Surg Oncol 23(11):3487–3493. https://doi.org/10.1245/s10434-016-5268-2
Nagi C, Guttman M, Jaffer S, Qiao R, Keren R, Triana A, Li M, Godbold J, Bleiweiss IJ, Hazan RB (2005) N-cadherin expression in breast cancer: correlation with an aggressive histologic variant–invasive micropapillary carcinoma. Breast Cancer Res Treat 94(3):225–235. https://doi.org/10.1007/s10549-005-7727-5
Aleskandarany MA, Soria D, Green AR, Nolan C, Diez-Rodriguez M, Ellis IO, Rakha EA (2015) Markers of progression in early-stage invasive breast cancer: a predictive immunohistochemical panel algorithm for distant recurrence risk stratification. Breast Cancer Res Treat 151(2):325–333. https://doi.org/10.1007/s10549-015-3406-3
Saadatmand S, de Kruijf EM, Sajet A, Dekker-Ensink NG, van Nes JG, Putter H, Smit VT, van de Velde CJ, Liefers GJ, Kuppen PJ (2013) Expression of cell adhesion molecules and prognosis in breast cancer. Br J Surg 100(2):252–260. https://doi.org/10.1002/bjs.8980
Bock C, Kuhn C, Ditsch N, Krebold R, Heublein S, Mayr D, Doisneau-Sixou S, Jeschke U (2014) Strong correlation between N-cadherin and CD133 in breast cancer: role of both markers in metastatic events. J Cancer Res Clin Oncol 140(11):1873–1881. https://doi.org/10.1007/s00432-014-1750-z
Hanna WM, Parra-Herran C, Lu FI, Slodkowska E, Rakovitch E, Nofech-Mozes S (2019) Ductal carcinoma in situ of the breast: an update for the pathologist in the era of individualized risk assessment and tailored therapies. Mod Pathol. https://doi.org/10.1038/s41379-019-0204-1
Maeda M, Johnson KR, Wheelock MJ (2005) Cadherin switching: essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J Cell Sci 118(Pt 5):873–887. https://doi.org/10.1242/jcs.01634
Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139(5):871–890. https://doi.org/10.1016/j.cell.2009.11.007
Qian X, Anzovino A, Kim S, Suyama K, Yao J, Hulit J, Agiostratidou G, Chandiramani N, McDaid HM, Nagi C, Cohen HW, Phillips GR, Norton L, Hazan RB (2014) N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene 33(26):3411–3421. https://doi.org/10.1038/onc.2013.310
Estecio MR, Issa JP (2011) Dissecting DNA hypermethylation in cancer. FEBS Lett 585(13):2078–2086. https://doi.org/10.1016/j.febslet.2010.12.001
Ehrlich M (2002) DNA methylation in cancer: too much, but also too little. Oncogene 21(35):5400–5413. https://doi.org/10.1038/sj.onc.1205651
Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I (2006) Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 38(2):149–153. https://doi.org/10.1038/ng1719
Sproul D, Nestor C, Culley J, Dickson JH, Dixon JM, Harrison DJ, Meehan RR, Sims AH, Ramsahoye BH (2011) Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proc Natl Acad Sci U S A 108(11):4364–4369. https://doi.org/10.1073/pnas.1013224108
Sproul D, Meehan RR (2013) Genomic insights into cancer-associated aberrant CpG island hypermethylation. Brief Funct Genomics 12(3):174–190. https://doi.org/10.1093/bfgp/els063
Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, Nitta KR, Taipale M, Popov A, Ginno PA, Domcke S, Yan J, Schubeler D, Vinson C, Taipale J (2017) Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. https://doi.org/10.1126/science.aaj2239
Zhang J, Brewer S, Huang J, Williams T (2003) Overexpression of transcription factor AP-2alpha suppresses mammary gland growth and morphogenesis. Dev Biol 256(1):127–145
Ailan H, Xiangwen X, Daolong R, Lu G, Xiaofeng D, Xi Q, Xingwang H, Rushi L, Jian Z, Shuanglin X (2009) Identification of target genes of transcription factor activator protein 2 gamma in breast cancer cells. BMC Cancer 9:279. https://doi.org/10.1186/1471-2407-9-279
Mrozik KM, Blaschuk OW, Cheong CM, Zannettino ACW, Vandyke K (2018) N-cadherin in cancer metastasis, its emerging role in haematological malignancies and potential as a therapeutic target in cancer. BMC Cancer 18(1):939. https://doi.org/10.1186/s12885-018-4845-0
Blaschuk OW (2015) N-cadherin antagonists as oncology therapeutics. Philos Trans R Soc Lond B Biol Sci 370(1661):20140039. https://doi.org/10.1098/rstb.2014.0039
Bui MM, Riben MW, Allison KH, Chlipala E, Colasacco C, Kahn AG, Lacchetti C, Madabhushi A, Pantanowitz L, Salama ME, Stewart RL, Thomas NE, Tomaszewski JE, Hammond ME (2019) Quantitative Image Analysis of Human Epidermal Growth Factor Receptor 2 Immunohistochemistry for Breast Cancer: Guideline From the College of American Pathologists. Arch Pathol Lab Med. https://doi.org/10.5858/arpa.2018-0378-CP
Acknowledgements
Biospecimens were provided by the National Cancer Institute-funded Cooperative Human Tissue Network, Tumor Tissue and Biospecimen Bank. and the Department of Pathology and Laboratory Medicine at the University of Pennsylvania. We are grateful to Caitlin Feltcher, Federico Valdivieso, Natalie Shih, Dee McGarvey, Deneen Mack, and Nicole Bollinger, and Gina Piermatteo for assistance with human tissue procurement and Li Ping for help with automated IHC. We thank Samantha Steeman for excellent assistance with editing and proofreading of this manuscript.
Funding
The authors received no external funding in support of this research.
Author information
Authors and Affiliations
Contributions
M.A.G. designed and supervised all aspects of this study. I.P. acquired samples and performed QG2 assay and gDNA extraction. Z.W. performed statistical modeling. D.I.F., Y.H., J.T. reviewed biopsy charts. P.J.Z. re-reviewed archival tissue slides and pathology reports. I.P. constructed databases. M.A.G. and Z.W. analyzed the data, M.A.G. and I.P. wrote the paper. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors. An approval from the University of Pennsylvania Institutional Review Board committee with a waiver of written informed consent was used for analysis of patients’ tissue and records.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2020_5797_MOESM3_ESM.pdf
Supplementary file3 Negative and positive tissue controls IHC –stained with an N –cadherin monoclonal antibody. Arrows, examples of microvasculature stained positive in IBC. Scale bar, 20µm. (PDF 31113 kb)
10549_2020_5797_MOESM4_ESM.pdf
Supplementary file4 Flow chart of inclusion and exclusion criteria of the study samples. *, preserved for the future patient care use; **, 5 years after treatment. HG, high grade, nonHG, non-high grade. (PDF 54 kb)
10549_2020_5797_MOESM5_ESM.pdf
Supplementary file5 Figure S3. Classification of CDH2-positive and CDH2-negative biopsy using GMM-based cutoff with confidence. A. GMM for overall (orange), similar to normal (green), and upregulated (red) gene expression in cancer are superimposed on histograms of frequency of CDH2 expression data in patients diagnosed with invasive breast cancer. B. Box plot depicts control CDH2 measurements in mammoplasties. Vertical lines in boxes, medians. Boxes. 25th and 75th percentile. Whiskers, 75th +1.5xIQR and 25th-1.5xIQR. X axis in A-C, mRNA levels in relative units. C. Graph for confidence calculated for invasive breast groups with changed gene expression as defined in Materials and Methods (blue). Step function plotted to illustrate minimal misclassification cutoffs as intersections of confidence and step function (green). The derivative of confidence with a half-height width plotted to define a range where the probability for sample to belong to either normal or changed group is similar (orange). The cutoff range was computed through the differentiation of confidence function followed by the calculation of the half-height width. D. Scatter plot for CDH2mRNA in the groups of mammoplasties (N, n=34), pure DCIS (DCIS, n=54), synchronous DCIS (DCIS w. INV, n=21), and IBC (n=133). E. Sample classification in normal (N), DCIS (DCIS) and DCIS with invasion (DCIS w. INV) groups based on three cutoffs. CDH2 positive and negative samples are depicted in red and green, respectively. The histogram, a summary of the proportion of CDH2 positive samples in each group classified with three cutoffs. (PDF 329 kb)
10549_2020_5797_MOESM6_ESM.pdf
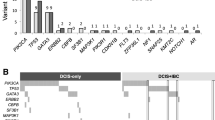
Supplementary file6 The analysis of fourteen ROC models predicting high-grade DCIS. Forest plots; Y axis, 14 genes; X axis, AUC, bar, 95% CI (PDF 34 kb)
10549_2020_5797_MOESM7_ESM.pdf
Supplementary file7 A. The methylation ratios (Y axis = 0-1) were compared in 157 individual CpG site between reference group (CDH2-, CDH2mRNA levels ≤2.2; n=12) and test group (CDH2+, CDH2mRNA levels >2.2; n=12) of DCIS. Samples were coded and examined in a blind fashion. Arrow, CpG_25757619 site affected by hypermethylation. B. Comparison summary of the methylation differences at each of 157 examined CpG site between the test and reference groups of DCIS. A site with statistically significant difference in the methylation is in red. P-value, Student’s test. (PDF 156 kb)
Rights and permissions
About this article
Cite this article
Guvakova, M.A., Prabakaran, I., Wu, Z. et al. CDH2/N-cadherin and early diagnosis of invasion in patients with ductal carcinoma in situ. Breast Cancer Res Treat 183, 333–346 (2020). https://doi.org/10.1007/s10549-020-05797-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-020-05797-x