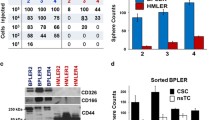
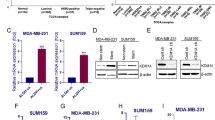
Abstract
Purpose
Breast cancer stem cells (CSCs) are a small subpopulation of cancer cells that have high capability for self-renewal, differentiation, and tumor initiation. CSCs are resistant to chemotherapy and radiotherapy, and are responsible for cancer recurrence and metastasis.
Methods
By utilizing a panel of breast cancer cells and mammospheres culture as cell-based screening platforms, we performed high-throughput chemical library screens to identify agents that are effective against breast CSCs and non-CSCs. The hit molecules were paired with conventional chemotherapy to evaluate the combinatorial treatment effects on breast CSCs and non-CSCs.
Results
We identified a total of 193 inhibitors that effectively targeting both breast CSCs and non-CSCs. We observed that histone deacetylase inhibitors (HDACi) synergized conventional chemotherapeutic agents (i.e., doxorubicin and cisplatin) in targeting breast CSCs and non-CSCs simultaneously. Further analyses revealed that quisinostat, a potent inhibitor for class I and II HDACs, potentiated doxorubicin-induced cytotoxicity in both breast CSCs and non-CSCs derived from the basal-like (MDA-MB-468 and HCC38), mesenchymal-like (MDA-MB-231), and luminal-like breast cancer (MCF-7). It was also observed that the basal-like breast CSCs and non-CSCs were more sensitive to the co-treatment of quisinostat with doxorubicin compared to that of the luminal-like breast cancer subtype.
Conclusion
In conclusion, this study demonstrates the potential of HDACi as therapeutic options, either as monotherapy or in combination with chemotherapeutics against refractory breast cancer.





Similar content being viewed by others
References
Batlle E, Clevers H (2017) Cancer stem cells revisited. Nat Med 23(10):1124–1134. https://doi.org/10.1038/nm.4409
Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY, Yaswen P, Goga A, Werb Z (2015) Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 526(7571):131–135. https://doi.org/10.1038/nature15260
Zhu Y, Zhang X, Liu Y, Zhang S, Liu J, Ma Y, Zhang J (2012) Antitumor effect of the mTOR inhibitor everolimus in combination with trastuzumab on human breast cancer stem cells in vitro and in vivo. Tumor Biol 33(5):1349–1362
Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, Margolick JB, Liotta LA, Petricoin E, Zhang Y (2007) Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci 104(41):16158–16163
Boyer M, Cheng T (2008) The CDK inhibitors: potential targets for therapeutic stem cell manipulations? Gene Ther 15(2):117
Han YK, Lee JH, Park G-Y, Chun SH, Han JY, Kim SD, Lee J, Lee C-W, Yang K, Lee CG (2013) A possible usage of a CDK4 inhibitor for breast cancer stem cell-targeted therapy. Biochem Biophys Res Commun 430(4):1329–1333
Chang W-W, Lin R-J, Yu J, Chang W-Y, Fu C-H, Lai AC-Y, Yu J-C, Alice LY (2013) The expression and significance of insulin-like growth factor-1 receptor and its pathway on breast cancer stem/progenitors. Breast Cancer Res 15(3):R39
Christopoulos PF, Msaouel P, Koutsilieris M (2015) The role of the insulin-like growth factor-1 system in breast cancer. Mol Cancer Res 14(1):1
Hormones TE, Group BCC (2010) Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol 11(6):530
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100(7):3983–3988. https://doi.org/10.1073/pnas.0530291100
Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS (2009) Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res 69(4):1302–1313. https://doi.org/10.1158/0008-5472.CAN-08-2741
Fillmore CM, Kuperwasser C (2008) Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res BCR 10(2):R25. https://doi.org/10.1186/bcr1982
Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG (2005) Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 65(13):5506–5511. https://doi.org/10.1158/0008-5472.CAN-05-0626
Soo JS, Ng CH, Tan SH, Malik RA, Teh YC, Tan BS, Ho GF, See MH, Taib NA, Yip CH, Chung FF, Hii LW, Teo SH, Leong CO (2015) Metformin synergizes 5-fluorouracil, epirubicin, and cyclophosphamide (FEC) combination therapy through impairing intracellular ATP production and DNA repair in breast cancer stem cells. Apoptosis 20(10):1373–1387. https://doi.org/10.1007/s10495-015-1158-5
Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K (2009) Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res 69(19):7507–7511. https://doi.org/10.1158/0008-5472.CAN-09-2994
Vazquez-Martin A, Oliveras-Ferraros C, Del Barco S, Martin-Castillo B, Menendez JA (2011) The anti-diabetic drug metformin suppresses self-renewal and proliferation of trastuzumab-resistant tumor-initiating breast cancer stem cells. Breast Cancer Res Treat 126(2):355–364. https://doi.org/10.1007/s10549-010-0924-x
Janzer A, German NJ, Gonzalez-Herrera KN, Asara JM, Haigis MC, Struhl K (2014) Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc Natl Acad Sci USA 111(29):10574–10579. https://doi.org/10.1073/pnas.1409844111
Iliopoulos D, Hirsch HA, Struhl K (2011) Metformin decreases the dose of chemotherapy for prolonging tumor remission in mouse xenografts involving multiple cancer cell types. Cancer Res 71(9):3196–3201. https://doi.org/10.1158/0008-5472.CAN-10-3471
Remsik J, Fedr R, Navratil J, Bino L, Slabakova E, Fabian P, Svoboda M, Soucek K (2018) Plasticity and intratumoural heterogeneity of cell surface antigen expression in breast cancer. Br J Cancer 118(6):813–819. https://doi.org/10.1038/bjc.2017.497
Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES (2009) Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138(4):645–659. https://doi.org/10.1016/j.cell.2009.06.034
Shibue T, Weinberg RA (2017) EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 14(10):611–629. https://doi.org/10.1038/nrclinonc.2017.44
Soo HC, Chung FF, Lim KH, Yap VA, Bradshaw TD, Hii LW, Tan SH, See SJ, Tan YF, Leong CO, Mai CW (2017) Cudraflavone C Induces Tumor-Specific Apoptosis in Colorectal Cancer Cells through Inhibition of the Phosphoinositide 3-Kinase (PI3 K)-AKT Pathway. PLoS ONE 12(1):e0170551. https://doi.org/10.1371/journal.pone.0170551
König R, Chiang C-y, Tu BP, Yan SF, DeJesus PD, Romero A, Bergauer T, Orth A, Krueger U, Zhou Y (2007) A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods 4(10):847
Tiong KH, Tan BS, Choo HL, Chung FF, Hii LW, Tan SH, Khor NT, Wong SF, See SJ, Tan YF, Rosli R, Cheong SK, Leong CO (2016) Fibroblast growth factor receptor 4 (FGFR4) and fibroblast growth factor 19 (FGF19) autocrine enhance breast cancer cells survival. Oncotarget 7(36):57633–57650. https://doi.org/10.18632/oncotarget.9328
Chung FF, Tan PF, Raja VJ, Tan BS, Lim KH, Kam TS, Hii LW, Tan SH, See SJ, Tan YF, Wong LZ, Yam WK, Mai CW, Bradshaw TD, Leong CO (2017) Jerantinine A induces tumor-specific cell death through modulation of splicing factor 3b subunit 1 (SF3B1). Scientific Rep 7:42504. https://doi.org/10.1038/srep42504
Stone EL, Citossi F, Singh R, Kaur B, Gaskell M, Farmer PB, Monks A, Hose C, Stevens MF, Leong CO, Stocks M, Kellam B, Marlow M, Bradshaw TD (2015) Antitumour benzothiazoles. Part 32: DNA adducts and double strand breaks correlate with activity; synthesis of 5F203 hydrogels for local delivery. Bioorg Med Chem 23(21):6891–6899. https://doi.org/10.1016/j.bmc.2015.09.052
Chou T-C (2010) Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70(2):440–446
Er JL, Goh PN, Lee CY, Tan YJ, Hii L-W, Mai CW, Chung FF-L, Leong C-O (2018) Identification of inhibitors synergizing gemcitabine sensitivity in the squamous subtype of pancreatic ductal adenocarcinoma (PDAC). Apoptosis 23:1–13
Voon YL, Ahmad M, Wong PF, Husaini R, Ng WT, Leong CO, Lane DP, Khoo AS (2015) Nutlin-3 sensitizes nasopharyngeal carcinoma cells to cisplatin-induced cytotoxicity. Oncol Rep 34(4):1692–1700. https://doi.org/10.3892/or.2015.4177
Low SY, Tan BS, Choo HL, Tiong KH, Khoo AS-B, Leong C-O (2012) Suppression of BCL-2 synergizes cisplatin sensitivity in nasopharyngeal carcinoma cells. Cancer Lett 314(2):166–175
Wong SW, Tiong KH, Kong WY, Yue YC, Chua CH, Lim JY, Lee CY, Quah SI, Fow C, Chung C (2011) Rapamycin synergizes cisplatin sensitivity in basal-like breast cancer cells through up-regulation of p73. Breast Cancer Res Treat 128(2):301–313
Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, Jodrell DI (2016) Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 32(18):2866–2868
Dai X, Cheng H, Bai Z, Li J (2017) Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer 8(16):3131–3141. https://doi.org/10.7150/jca.18457
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10(6):515–527. https://doi.org/10.1016/j.ccr.2006.10.008
Liu K, Newbury PA, Glicksberg BS, Zeng WZD, Paithankar S, Andrechek ER, Chen B (2019) Evaluating cell lines as models for metastatic breast cancer through integrative analysis of genomic data. Nat Commun 10(1):2138. https://doi.org/10.1038/s41467-019-10148-6
Li W, Ma H, Zhang J, Zhu L, Wang C, Yang Y (2017) Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Scientific Rep 7(1):13856. https://doi.org/10.1038/s41598-017-14364-2
Fillmore CM, Kuperwasser C (2008) Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 10(2):R25
Venere M, Horbinski C, Crish JF, Jin X, Vasanji A, Major J, Burrows AC, Chang C, Prokop J, Wu Q, Sims PA, Canoll P, Summers MK, Rosenfeld SS, Rich JN (2015) The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med 7(304):304ra143. https://doi.org/10.1126/scitranslmed.aac6762
Cheng CC, Chang J, Huang SC, Lin HC, Ho AS, Lim KH, Chang CC, Huang L, Chang YC, Chang YF, Wu CW (2017) YM155 as an inhibitor of cancer stemness simultaneously inhibits autophosphorylation of epidermal growth factor receptor and G9a-mediated stemness in lung cancer cells. PLoS ONE 12(8):e0182149. https://doi.org/10.1371/journal.pone.0182149
Vequaud E, Seveno C, Loussouarn D, Engelhart L, Campone M, Juin P, Barille-Nion S (2015) YM155 potently triggers cell death in breast cancer cells through an autophagy-NF-kB network. Oncotarget 6(15):13476–13486. https://doi.org/10.18632/oncotarget.3638
Huang M, Liu B, Liu R, Li J, Chen J, Jiang F, Ding H, Deng Z, Liu T (2018) Aglycone Polyether Nanchangmycin and Its Homologues Exhibit Apoptotic and Antiproliferative Activities against Cancer Stem Cells. ACS Pharmacol Transl Sci 1(2):84–95. https://doi.org/10.1021/acsptsci.8b00007
Yunokawa M, Koizumi F, Kitamura Y, Katanasaka Y, Okamoto N, Kodaira M, Yonemori K, Shimizu C, Ando M, Masutomi K (2012) Efficacy of everolimus, a novel m TOR inhibitor, against basal-like triple-negative breast cancer cells. Cancer Sci 103(9):1665–1671
Wander SA, Hennessy BT, Slingerland JM (2011) Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Investig 121(4):1231–1241
Kretsovali A, Hadjimichael C, Charmpilas N (2012) Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem cells international 2012:10
Bolden JE, Peart MJ, Johnstone RW (2006) Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 5(9):769–784. https://doi.org/10.1038/nrd2133
West AC, Johnstone RW (2014) New and emerging HDAC inhibitors for cancer treatment. J Clin Investig 124(1):30–39. https://doi.org/10.1172/JCI69738
Ceccacci E, Minucci S (2016) Inhibition of histone deacetylases in cancer therapy: lessons from leukaemia. Br J Cancer 114(6):605
Chiappinelli KB, Zahnow CA, Ahuja N, Baylin SB (2016) Combining epigenetic and immunotherapy to combat cancer. Cancer Res 76(7):1683–1689. https://doi.org/10.1158/0008-5472.CAN-15-2125
Venugopal B, Baird R, Kristeleit RS, Plummer R, Cowan R, Stewart A, Fourneau N, Hellemans P, Elsayed Y, McClue S, Smit JW, Forslund A, Phelps C, Camm J, Evans TR, de Bono JS, Banerji U (2013) A phase I study of quisinostat (JNJ-26481585), an oral hydroxamate histone deacetylase inhibitor with evidence of target modulation and antitumor activity, in patients with advanced solid tumors. Clin Cancer Res 19(15):4262–4272. https://doi.org/10.1158/1078-0432.CCR-13-0312
He B, Dai L, Zhang X, Chen D, Wu J, Feng X, Zhang Y, Xie H, Zhou L, Zheng S (2018) The HDAC Inhibitor Quisinostat (JNJ-26481585) Supresses Hepatocellular Carcinoma alone and Synergistically in Combination with Sorafenib by G0/G1 phase arrest and Apoptosis induction. Int J Biol Sci 14(13):1845–1858. https://doi.org/10.7150/ijbs.27661
Tjulandin S, Fedyanin M, Vladimirov VI, Kostorov V, Lisyanskaya AS, Krikunova L, Cakana A, Azarova V, Karavaeva O, Vostokova N (2017) A multicenter phase II study of the efficacy and safety of quisinostat (an HDAC inhibitor) in combination with paclitaxel and carboplatin chemotherapy (CT) in patients (pts) with recurrent platinum resistant high grade serous epithelial ovarian, primarily peritoneal or fallopian tube carcinoma cancer (OC). American Society of Clinical Oncology,
Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF (2003) Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci 983(1):84–100
Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M (2012) Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 11(5):384–400. https://doi.org/10.1038/nrd3674
Müller BM, Jana L, Kasajima A, Lehmann A, Prinzler J, Budczies J, Winzer K-J, Dietel M, Weichert W, Denkert C (2013) Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer-overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer 13(1):215
Seo J, Min SK, Park H-R, Kim DH, Kwon MJ, Kim LS, Ju Y-S (2014) Expression of histone deacetylases HDAC1, HDAC2, HDAC3, and HDAC6 in invasive ductal carcinomas of the breast. J Breast Cancer 17(4):323–331
Witt AE, Lee CW, Lee TI, Azzam DJ, Wang B, Caslini C, Petrocca F, Grosso J, Jones M, Cohick EB, Gropper AB, Wahlestedt C, Richardson AL, Shiekhattar R, Young RA, Ince TA (2017) Identification of a cancer stem cell-specific function for the histone deacetylases, HDAC1 and HDAC7, in breast and ovarian cancer. Oncogene 36(12):1707–1720. https://doi.org/10.1038/onc.2016.337
Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H (2005) Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast. Breast Cancer Res Treat 94(1):11–16
Glaser KB, Li J, Staver MJ, Wei R-Q, Albert DH, Davidsen SK (2003) Role of class I and class II histone deacetylases in carcinoma cells using siRNA. Biochem Biophys Res Commun 310(2):529–536
Rey M, Irondelle M, Waharte F, Lizarraga F, Chavrier P (2011) HDAC6 is required for invadopodia activity and invasion by breast tumor cells. Eur J Cell Biol 90(2–3):128–135
Delcuve GP, Khan DH, Davie JR (2012) Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenet 4(1):5
Falkenberg KJ, Johnstone RW (2014) Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 13(9):673
Guzman ML, Yang N, Sharma KK, Balys M, Corbett CA, Jordan CT, Becker MW, Steidl U, Abdel-Wahab O, Levine RL, Marcucci G, Roboz GJ, Hassane DC (2014) Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: a novel potential strategy in acute myelogenous leukemia. Mol Cancer Ther 13(8):1979–1990. https://doi.org/10.1158/1535-7163.MCT-13-0963
Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S (2011) Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS ONE 6(8):e24099. https://doi.org/10.1371/journal.pone.0024099
Kumar B, Yadav A, Lang JC, Teknos TN, Kumar P (2015) Suberoylanilide hydroxamic acid (SAHA) reverses chemoresistance in head and neck cancer cells by targeting cancer stem cells via the downregulation of nanog. Genes Cancer 6(3–4):169–181. https://doi.org/10.18632/genesandcancer.54
Salvador MA, Wicinski J, Cabaud O, Toiron Y, Finetti P, Josselin E, Lelievre H, Kraus-Berthier L, Depil S, Bertucci F, Collette Y, Birnbaum D, Charafe-Jauffret E, Ginestier C (2013) The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin Cancer Res 19(23):6520–6531. https://doi.org/10.1158/1078-0432.CCR-13-0877
Di Pompo G, Salerno M, Rotili D, Valente S, Zwergel C, Avnet S, Lattanzi G, Baldini N, Mai A (2015) Novel histone deacetylase inhibitors induce growth arrest, apoptosis, and differentiation in sarcoma cancer stem cells. J Med Chem 58(9):4073–4079. https://doi.org/10.1021/acs.jmedchem.5b00126
Seto E, Yoshida M (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6(4):a018713. https://doi.org/10.1101/cshperspect.a018713
Liu N, Li S, Wu N, Cho KS (2017) Acetylation and deacetylation in cancer stem-like cells. Oncotarget 8(51):89315–89325. https://doi.org/10.18632/oncotarget.19167
Doherty MR, Smigiel JM, Junk DJ, Jackson MW (2016) Cancer stem cell plasticity drives therapeutic resistance. Cancers 8(1):E8. https://doi.org/10.3390/cancers8010008
Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, Weinberg RA (2015) Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525(7568):256–260. https://doi.org/10.1038/nature14897
Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, Arendt LM, Kuperwasser C, Bierie B, Weinberg RA (2011) Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA 108(19):7950–7955. https://doi.org/10.1073/pnas.1102454108
Ravikumar B, Alam Z, Peddinti G, Aittokallio T (2017) C-SPADE: a web-tool for interactive analysis and visualization of drug screening experiments through compound-specific bioactivity dendrograms. Nucleic Acids Res 45(W1):W495–W500. https://doi.org/10.1093/nar/gkx384
Funding
This study was funded by the Malaysia Ministry of Education Exploratory Research Grant Scheme (LCO; ERGS/1/2013/SKK01/IMU/02/1) and Malaysia Ministry of Education Fundamental Grant Scheme (LCO; FRGS/1/2016/SKK08/IMU/01/1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2019_5504_MOESM2_ESM.pptx
Supplementary material 2 (PPTX 1110 kb). Supplementary Figure 1: Breast CSCs are intrinsically resistant to conventional chemotherapeutic agents. Both CSCs and non-CSCs derived from MDA-MB-468, MDA-MB-231, HCC38, and MCF-7 breast cancer cell lines were treated with cisplatin, doxorubicin and paclitaxel for 72 h. Points represent mean ± S.D. of at least three independent experiments. Supplemental Figure 2: Combinatory effects of HDACi and paclitaxel on MDA-MB-468 breast CSCs and non-CSCs. MDA-MB-468 breast CSCs and non-CSCs were treated with paclitaxel and/or HDACi for 72 h. Dose–response surface curves and synergy of each combination was assessed using the HSA model (effect-based approach), as implemented in Combenefit software [32]. Level of synergism (blue) or antagonism (red) at each concentration is represented by color scale bar. All experiments were conducted at least three times
Rights and permissions
About this article
Cite this article
Hii, LW., Chung, F.FL., Soo, J.SS. et al. Histone deacetylase (HDAC) inhibitors and doxorubicin combinations target both breast cancer stem cells and non-stem breast cancer cells simultaneously. Breast Cancer Res Treat 179, 615–629 (2020). https://doi.org/10.1007/s10549-019-05504-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-019-05504-5