Abstract
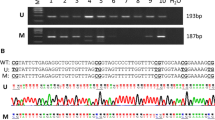
BRCAness breast tumors represent a group of sporadic tumors characterized by a reduction in BRCA1 gene expression. As BRCA1 is involved in double-strand breaks (DSBs) repair, dysfunctional BRCA pathway could make a tumor sensitive to DNA damaging drugs (e.g., platinum agents). Thus, accurately identifying BRCAness could contribute to therapeutic decision making in patients harboring these tumors. The purpose of this study was to identify if BRCAness tumors present a characteristic methylation profile and/or were related to specific clinico-pathological features. BRCAness was measured by MLPA in 63 breast tumors; methylation status of 98 CpG sites within 84 cancer-related genes was analyzed by MS-MLPA. Protein and mRNA expressions of the selected genes were measured by quantitative real-time PCR and Western Blot. BRCAness was associated with younger age, higher nuclear pleomorfism, and triple-negative (TN) status. Epigenetically, we found that the strongest predictors for BRCAness tumors were the methylations of MLH1 and PAX5 plus the unmethylations of CCND2 and ID4. We determined that ID4 unmethylation correlated with the expression levels of both its mRNA and protein. We observed an inverse relation between the expressions of ID4 and BRCA1. To the best of our knowledge, this is the first report suggesting an epigenetic regulation of ID4 in BRCAness tumors. Our findings give new information of BRCAness etiology and encourage future studies on potential drug targets for BRCAness breast tumors.





Similar content being viewed by others
References
Larsen MJ, Thomassen M, Gerdes AM, Kruse TA (2014) Hereditary breast cancer: clinical, pathological and molecular characteristics. Breast Cancer (Auckl.) 8:145–155
Domchek SM (2015) Evolution of genetic testing for inherited susceptibility to breast cancer. J Clin Oncol 33:295–296
Turner N, Tutt A, Ashworth A (2004) Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 4:814–819
Brekelmans CT, Seynaeve C, Menke-Pluymers M, Bruggenwirth HT, Tilanus-Linthorst MM, Bartels CC, Kriege M, Geel AN, Crepin CM, Blom JC, Meijers-Heijboer H, Klijn JG (2006) Survival and prognostic factors in BRCA1-associated breast cancer. Ann Oncol 17:391–400
Arnes JB, Brunet JS, Stefansson I, Begin LR, Wong N, Chappuis PO, Akslen LA, Foulkes WD (2005) Placental cadherin and the basal epithelial phenotype of BRCA1-related breast cancer. Clin Cancer Res 11:4003–4011
Apostolou P, Fostira F (2013) Hereditary breast cancer: the era of new susceptibility genes. Biomed Res Int 2013:747318
Chalasani P, Livingston R (2013) Differential chemotherapeutic sensitivity for breast tumors with “BRCAness”: a review. Oncologist 18:909–916
Friedenson B (2005) BRCA1 and BRCA2 pathways and the risk of cancers other than breast or ovarian. MedGenMed 7:60
Carey JP, Knowell AE, Chinaranagari S, Chaudhary J (2013) Id4 promotes senescence and sensitivity to doxorubicin-induced apoptosis in DU145 prostate cancer cells. Anticancer Res 33:4271–4278
Fontemaggi G, Dell’Orso S, Trisciuoglio D, Shay T, Melucci E, Fazi F, Terrenato I, Mottolese M, Muti P, Domany E, Del BD, Strano S, Blandino G (2009) The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat Struct Mol Biol 16:1086–1093
Galatro TF, Uno M, Oba-Shinjo SM, Almeida AN, Teixeira MJ, Rosemberg S, Marie SK (2013) Differential expression of ID4 and its association with TP53 mutation, SOX2, SOX4 and OCT-4 expression levels. PLoS ONE 8:e61605
Umetani N, Mori T, Koyanagi K, Shinozaki M, Kim J, Giuliano AE, Hoon DS (2005) Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene 24:4721–4727
Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, Welch PJ, Welte K, King MC, Barber JR, Wong-Staal F (2001) Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA 98:130–135
Branham MT, Marzese DM, Laurito SR, Gago FE, Orozco JI, Tello OM, Vargas-Roig LM, Roque M (2012) Methylation profile of triple-negative breast carcinomas. Oncogenesis 1:e17
Marzese DM, Gago FE, Vargas-Roig LM, Roque M (2010) Simultaneous analysis of the methylation profile of 26 cancer related regions in invasive breast carcinomas by MS-MLPA and drMS-MLPA. Mol Cell Probes 24:271–280
Lips EH, Laddach N, Savola SP, Vollebergh MA, Oonk AM, Imholz AL, Wessels LF, Wesseling J, Nederlof PM, Rodenhuis S (2011) Quantitative copy number analysis by multiplex ligation-dependent probe amplification (MLPA) of BRCA1-associated breast cancer regions identifies BRCAness. Breast Cancer Res 13:R107
Joosse SA, Beers EH, Tielen IH, Horlings H, Peterse JL, Hoogerbrugge N, Ligtenberg MJ, Wessels LF, Axwijk P, Verhoef S, Hogervorst FB, Nederlof PM (2009) Prediction of BRCA1-association in hereditary non-BRCA1/2 breast carcinomas with array-CGH. Breast Cancer Res Treat 116:479–489
Oonk AM, van Rijn C, Smits MM, Mulder L, Laddach N, Savola SP, Wesseling J, Rodenhuis S, Imholz AL, Lips EH (2012) Clinical correlates of ‘BRCAness’ in triple-negative breast cancer of patients receiving adjuvant chemotherapy. Ann Oncol 23:2301–2305
Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A (2005) Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res 33:e128
Marzese DM, Gago FE, Vargas-Roig LM, Roque M (2010) Simultaneous analysis of the methylation profile of 26 cancer related regions in invasive breast carcinomas by MS-MLPA and drMS-MLPA. Mol Cell Probes 24:271–280
Urrutia G, Laurito S, Marzese DM, Gago F, Orozco J, Tello O, Branham T, Campoy EM, Roque M (2015) Epigenetic variations in breast cancer progression to lymph node metastasis. Clin Exp Metastasis 32:99–110
Lips EH, Mulder L, Oonk A, Kolk LE, Hogervorst FB, Imholz AL, Wesseling J, Rodenhuis S, Nederlof PM (2013) Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br J Cancer 108:2172–2177
Dell’Orso S, Ganci F, Strano S, Blandino G, Fontemaggi G (2010) ID4: a new player in the cancer arena. Oncotarget 1:48–58
Noetzel E, Veeck J, Niederacher D, Galm O, Horn F, Hartmann A, Knuchel R, Dahl E (2008) Promoter methylation-associated loss of ID4 expression is a marker of tumour recurrence in human breast cancer. BMC Cancer 8:154
Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De NA, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE (2010) Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol 28:1145–1153
Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, Savage K, Gillett CE, Schmitt FC, Ashworth A, Tutt AN (2007) BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 26:2126–2132
Suh DH, Kim MK, Kim HS, Chung HH, Song YS (2013) Epigenetic therapies as a promising strategy for overcoming chemoresistance in epithelial ovarian cancer. J. Cancer Prev 18:227–234
Lee JY, Jeong W, Kim JH, Kim J, Bazer FW, Han JY, Song G (2012) Distinct expression pattern and post-transcriptional regulation of cell cycle genes in the glandular epithelia of avian ovarian carcinomas. PLoS ONE 7:e51592
Moelans CB, Verschuur-Maes AH, van Diest PJ (2011) Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J Pathol 225:222–231
Crippa E, Lusa L, De CL, Marchesi E, Calin GA, Radice P, Manoukian S, Peissel B, Daidone MG, Gariboldi M, Pierotti MA (2014) miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PLoS ONE 9:e87039
Chatagnon A, Perriaud L, Nazaret N, Croze S, Benhattar J, Lachuer J, Dante R (2011) Preferential binding of the methyl-CpG binding domain protein 2 at methylated transcriptional start site regions. Epigenetics 6:1295–1307
Martini M, Cenci T, D’Alessandris GQ, Cesarini V, Cocomazzi A, Ricci-Vitiani L, De MR, Pallini R, Larocca LM (2013) Epigenetic silencing of Id4 identifies a glioblastoma subgroup with a better prognosis as a consequence of an inhibition of angiogenesis. Cancer 119:1004–1012
Roldan G, Delgado L, Muse IM (2006) Tumoral expression of BRCA1, estrogen receptor alpha and ID4 protein in patients with sporadic breast cancer. Cancer Biol Ther 5:505–510
de Candia P, Benera R, Solit DB (2004) A role for Id proteins in mammary gland physiology and tumorigenesis. Adv Cancer Res 92:81–94
Perk J, Iavarone A, Benezra R (2005) Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer 5:603–614
Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, Warrington JA, King MC (2002) BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci USA 99:7560–7565
Reinbolt RE, Hays JL (2013) The role of PARP inhibitors in the treatment of gynecologic malignancies. Front Oncol 3:237
Branham RL Jr (1990) Scientific data analysis. Springer, New York
Van Huffel S, Wandewalle J (1991) The total least squares problem: computational aspects and analysis. SIAM, Philadelphia
Branham RL Jr (2001) Astronomical data reduction with total least squares. New Astron Rev 45:649–661
Acknowledgments
This study was supported in part by the National Institute of Cancer (“Instituto Nacional del Cancer”), Argentina. RU was supported by funding from the National Institutes of Health (grant DK52913), the Mayo Clinic SPORE in Pancreatic Cancer (P50 CA102701), and the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567 to GL).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Human and animal rights
This article does not contain any studies with animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Branham, M.T., Campoy, E., Laurito, S. et al. Epigenetic regulation of ID4 in the determination of the BRCAness phenotype in breast cancer. Breast Cancer Res Treat 155, 13–23 (2016). https://doi.org/10.1007/s10549-015-3648-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-015-3648-0