Abstract
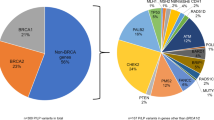
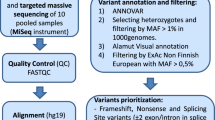
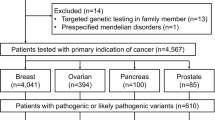
Multi-gene panels are used to identify genetic causes of hereditary breast and ovarian cancer (HBOC) in large patient cohorts. This study compares the diagnostic workflow in two centers and gives valuable insights into different next-generation sequencing (NGS) strategies. Moreover, we present data from 620 patients sequenced at both centers. Both sequencing centers are part of the German consortium for hereditary breast and ovarian cancer (GC-HBOC). All 620 patients included in this study were selected following standard BRCA1/2 testing guidelines. A set of 10 sequenced genes was analyzed per patient. Twelve samples were exchanged and sequenced at both centers. NGS results were highly concordant in 12 exchanged samples (205/206 variants = 99.51 %). One non-pathogenic variant was missed at center B due to a sequencing gap (no technical coverage). The custom enrichment at center B was optimized during this study; for example, the average number of missing bases was reduced by a factor of four (vers. 1: 1939.41, vers. 4: 506.01 bp). There were no sequencing gaps at center A, but four CCDS exons were not included in the enrichment. Pathogenic mutations were found in 12.10 % (75/620) of all patients: 4.84 % (30/620) in BRCA1, 4.35 % in BRCA2 (27/620), 0.97 % in CHEK2 (6/620), 0.65 % in ATM (4/620), 0.48 % in CDH1 (3/620), 0.32 % in PALB2 (2/620), 0.32 % in NBN (2/620), and 0.16 % in TP53 (1/620). NGS diagnostics for HBOC-related genes is robust, cost effective, and the method of choice for genetic testing in large cohorts. Adding 8 genes to standard BRCA1- and BRCA2-testing increased the mutation detection rate by one-third.



Similar content being viewed by others
References
Ferlay J, Parkin DM, Steliarova-Foucher E (2010) Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer 46(4):765–781. doi:10.1016/j.ejca.2009.12.014
Collaborative Group on Hormonal Factors in Breast C (2001) Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet 358(9291):1389–1399. doi:10.1016/S0140-6736(01)06524-2
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W et al (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266(5182):66–71
Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378(6559):789–792. doi:10.1038/378789a0
Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72(5):1117–1130. doi:10.1086/375033
Chen S, Iversen ES, Friebel T, Finkelstein D, Weber BL, Eisen A, Peterson LE, Schildkraut JM, Isaacs C, Peshkin BN, Corio C, Leondaridis L, Tomlinson G, Dutson D, Kerber R, Amos CI, Strong LC, Berry DA, Euhus DM, Parmigiani G (2006) Characterization of BRCA1 and BRCA2 mutations in a large United States sample. J Clin Oncol 24(6):863–871. doi:10.1200/JCO.2005.03.6772
Chen S, Parmigiani G (2007) Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol 25(11):1329–1333. doi:10.1200/JCO.2006.09.1066
Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Fan I, Tang J, Li S, Zhang S, Shaw PA, Narod SA (2006) Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst 98(23):1694–1706. doi:10.1093/jnci/djj465
Wooster R, Weber BL (2003) Breast and ovarian cancer. The New Engl J Med 348(23):2339–2347. doi:10.1056/NEJMra012284
Nathanson KL, Wooster R, Weber BL (2001) Breast cancer genetics: what we know and what we need. Nat Med 7(5):552–556. doi:10.1038/87876
Meindl A, German Consortium for Hereditary B, Ovarian C (2002) Comprehensive analysis of 989 patients with breast or ovarian cancer provides BRCA1 and BRCA2 mutation profiles and frequencies for the German population. Int J Cancer 97(4):472–480
Turnbull C, Rahman N (2008) Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet 9:321–345. doi:10.1146/annurev.genom.9.081307.164339
Apostolou P, Fostira F (2013) Hereditary breast cancer: the era of new susceptibility genes. BioMed Res Int 2013:747318. doi:10.1155/2013/747318
Bradbury AR, Olopade OI (2007) Genetic susceptibility to breast cancer. Rev Endocr Metabol Disord 8(3):255–267. doi:10.1007/s11154-007-9038-0
Ripperger T, Gadzicki D, Meindl A, Schlegelberger B (2009) Breast cancer susceptibility: current knowledge and implications for genetic counselling. Eur J Hum Genet 17(6):722–731. doi:10.1038/ejhg.2008.212
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11(1):31–46. doi:10.1038/nrg2626
Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ (2010) Target-enrichment strategies for next-generation sequencing. Nat Methods 7(2):111–118. doi:10.1038/nmeth.1419
Kurian AW, Kingham KE, Ford JM (2015) Next-generation sequencing for hereditary breast and gynecologic cancer risk assessment. Curr Opin Obstet Gynecol 27(1):23–33. doi:10.1097/GCO.0000000000000141
Wockel A, Kreienberg R (2008) First revision of the German S3 guideline ‘diagnosis, Therapy, and follow-up of breast cancer’. Breast care 3(2):82–86. doi:10.1159/000127509
Lunter G, Goodson M (2011) Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21(6):936–939. doi:10.1101/gr.111120.110
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. doi:10.1093/bioinformatics/btp698
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. doi:10.1093/bioinformatics/btp352
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43(5):491–498. doi:10.1038/ng.806
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164. doi:10.1093/nar/gkq603
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29(1):24–26. doi:10.1038/nbt.1754
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19(9):1639–1645. doi:10.1101/gr.092759.109
Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV, Group IUGVW (2008) Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 29(11):1282–1291. doi:10.1002/humu.20880
Sikkema-Raddatz B, Johansson LF, de Boer EN, Almomani R, Boven LG, van den Berg MP, van Spaendonck-Zwarts KY, van Tintelen JP, Sijmons RH, Jongbloed JD, Sinke RJ (2013) Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat 34(7):1035–1042. doi:10.1002/humu.22332
Cybulski C, Lubinski J, Wokolorczyk D, Kuzniak W, Kashyap A, Sopik V, Huzarski T, Gronwald J, Byrski T, Szwiec M, Jakubowska A, Gorski B, Debniak T, Narod SA, Akbari MR (2014) Mutations predisposing to breast cancer in 12 candidate genes in breast cancer patients from Poland. Clin Genet. doi:10.1111/cge.12524
Trujillano D, Weiss ME, Schneider J, Koster J, Papachristos EB, Saviouk V, Zakharkina T, Nahavandi N, Kovacevic L, Rolfs A (2015) Next-generation sequencing of the BRCA1 and BRCA2 genes for the genetic diagnostics of hereditary breast and/or ovarian cancer. J Mol Diagn 17(2):162–170. doi:10.1016/j.jmoldx.2014.11.004
Feliubadalo L, Lopez-Doriga A, Castellsague E, del Valle J, Menendez M, Tornero E, Montes E, Cuesta R, Gomez C, Campos O, Pineda M, Gonzalez S, Moreno V, Brunet J, Blanco I, Serra E, Capella G, Lazaro C (2013) Next-generation sequencing meets genetic diagnostics: development of a comprehensive workflow for the analysis of BRCA1 and BRCA2 genes. Eur J Hum Genet 21(8):864–870. doi:10.1038/ejhg.2012.270
Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, Freund M, Lichtner P, Hartmann L, Schaal H, Ramser J, Honisch E, Kubisch C, Wichmann HE, Kast K, Deissler H, Engel C, Muller-Myhsok B, Neveling K, Kiechle M, Mathew CG, Schindler D, Schmutzler RK, Hanenberg H (2010) Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 42(5):410–414. doi:10.1038/ng.569
Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, Bowden G, Kalmyrzaev B, Warren-Perry M, Snape K, Adlard JW, Barwell J, Berg J, Brady AF, Brewer C, Brice G, Chapman C, Cook J, Davidson R, Donaldson A, Douglas F, Greenhalgh L, Henderson A, Izatt L, Kumar A, Lalloo F, Miedzybrodzka Z, Morrison PJ, Paterson J, Porteous M, Rogers MT, Shanley S, Walker L, Breast Cancer Susceptibility C, Eccles D, Evans DG, Renwick A, Seal S, Lord CJ, Ashworth A, Reis-Filho JS, Antoniou AC, Rahman N (2011) Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet 43(9):879–882. doi:10.1038/ng.893
Maxwell KN, Wubbenhorst B, D’Andrea K, Garman B, Long JM, Powers J, Rathbun K, Stopfer JE, Zhu J, Bradbury AR, Simon MS, DeMichele A, Domchek SM, Nathanson KL (2014) Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genet Med. doi:10.1038/gim.2014.176
Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, Nord AS, Mandell JB, Swisher EM, King MC (2010) Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci USA 107(28):12629–12633. doi:10.1073/pnas.1007983107
Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, McGuire V, Ladabaum U, Kobayashi Y, Lincoln SE, Cargill M, Ford JM (2014) Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol 32(19):2001–2009. doi:10.1200/JCO.2013.53.6607
Chong HK, Wang T, Lu HM, Seidler S, Lu H, Keiles S, Chao EC, Stuenkel AJ, Li X, Elliott AM (2014) The validation and clinical implementation of BRCAplus: a comprehensive high-risk breast cancer diagnostic assay. PLoS ONE 9(5):e97408. doi:10.1371/journal.pone.0097408
Arvai K, Horvath P, Balla B, Tokes AM, Tobias B, Takacs I, Nagy Z, Lakatos P, Kosa JP (2014) Rapid and cost effective screening of breast and ovarian cancer genes using novel sequence capture method in clinical samples. Fam Cancer 13(4):583–589. doi:10.1007/s10689-014-9730-7
Castera L, Krieger S, Rousselin A, Legros A, Baumann JJ, Bruet O, Brault B, Fouillet R, Goardon N, Letac O, Baert-Desurmont S, Tinat J, Bera O, Dugast C, Berthet P, Polycarpe F, Layet V, Hardouin A, Frebourg T, Vaur D (2014) Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet 22(11):1305–1313. doi:10.1038/ejhg.2014.16
Tung N, Battelli C, Allen B, Kaldate R, Bhatnagar S, Bowles K, Timms K, Garber JE, Herold C, Ellisen L, Krejdovsky J, DeLeonardis K, Sedgwick K, Soltis K, Roa B, Wenstrup RJ, Hartman AR (2015) Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer 121(1):25–33. doi:10.1002/cncr.29010
Acknowledgments
The authors thank all patients and families for their participation in this study. This study was supported by the German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Schroeder, C., Faust, U., Sturm, M. et al. HBOC multi-gene panel testing: comparison of two sequencing centers. Breast Cancer Res Treat 152, 129–136 (2015). https://doi.org/10.1007/s10549-015-3429-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-015-3429-9