Abstract
Estrogen catabolism is a major function of CYP2C19. The effect of CYP2C19 polymorphisms on tamoxifen sensitivity may therefore not only be mediated by a variation in tamoxifen metabolite levels but also by an effect on breast cancer risk and molecular subtype due to variation in lifelong exposure to estrogens. We determined the association between these polymorphisms and tamoxifen sensitivity in the context of a randomized trial, which allows for the discernment of prognosis from prediction. We isolated primary tumor DNA from 535 estrogen receptor-positive, stages I–III, postmenopausal breast cancer patients who had been randomized to tamoxifen (1–3 years) or no adjuvant therapy. Recurrence-free interval improvement with tamoxifen versus control was assessed according to the presence or absence of CYP2C19*2 and CYP2C19*17. Hazard ratios and interaction terms were calculated using multivariate Cox proportional hazard models, stratified for nodal status. Tamoxifen benefit was not significantly affected by CYP2C19*17. Patients with at least one CYP2C19*2 allele derived significantly more benefit from tamoxifen (HR 0.26; p = 0.001) than patients without a CYP2C19*2 allele (HR 0.68; p = 0.18) (p for interaction 0.04). In control patients, CYP2C19*2 was an adverse prognostic factor. In conclusion, breast cancer patients carrying at least one CYP2C19*2 allele have an adverse prognosis in the absence of adjuvant systemic treatment, which can be substantially improved by adjuvant tamoxifen treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
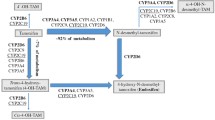
Although polymorphisms in CYP450 enzymes are extensively studied in the context of drug metabolism, the primary physiologic role of these enzymes is the metabolism of endogenous steroids, like catabolism of estrogens [1]. Thereby, in breast cancer patients, a potentially observed effect of a CYP450 polymorphism on tamoxifen sensitivity may not only be mediated by variation in tamoxifen metabolite levels but also possibly by variation in lifelong exposure to estrogens that affect tumorigenesis.
CYP2C19 is a typical example of a CYP450 enzyme that affects both metabolism of tamoxifen to 4-hydroxytamoxifen [2] as well as estrone (E1) and estradiol (E2) catabolism [1]. In addition, CYP2C19 has been shown to catalyze metabolism of testosterone [3]. Both estrogens and testosterone have been demonstrated to affect the risk of estrogen receptor α (ERα)-positive breast cancer in postmenopausal women [4]. Relatively frequent CYP2C19 polymorphisms are the (non-functional) CYP2C19*2 variant, with a minor allele frequency of 13 % in healthy Caucasians [5], and the (ultra-active) CYP2C19*17 variant, with a minor allele frequency of around 20 % [6]. Previously, it has been shown in postmenopausal breast cancer patients that genetic variation in CYP2C19 affects estrone levels [7]. The highest estrone levels were found in patients who were either heterozygous or homozygous for the CYP2C19*2 allele, while the ultrarapid variant of this enzyme, CYP2C19*17, was associated with low estrone levels [7] and decreased breast cancer risk [8].
Estrogens are not only associated with breast cancer risk but also possibly influence breast cancer molecular subtype [9], and therefore polymorphisms that affect estrogen levels might have an effect on breast cancer prognosis and drug sensitivity. Intriguingly, both the non-functional CYP2C19 variant as well as the ultrarapid variant have been associated with favorable outcomes after endocrine therapy. CYP2C19*2 was associated with a favorable progression-free survival in patients with metastatic breast cancer treated with tamoxifen [10], while carriers of a CYP2C19*17 allele who were treated with adjuvant tamoxifen had a favorable disease-free survival compared to non-CYP2C19*17 carriers [11]. A general methodological flaw in studies which analyze the polymorphism in consecutive series of patients treated with the same drug is that the predictive value of these polymorphisms with respect to drug sensitivity cannot be discriminated from the prognostic value. Therefore, the optimal method of discerning a predictive marker from a prognostic marker is within a randomized clinical trial.
The aim of our study was to analyze the predictive and prognostic value of genetic polymorphisms in CYP2C19 in postmenopausal breast cancer patients, randomized between adjuvant tamoxifen and no systemic treatment.
Methods
Patients and material
From 1982 to 1994, a randomized clinical trial was conducted in the Netherlands, studying the benefit from adjuvant tamoxifen (IKA-trial) versus no adjuvant therapy [12, 13]. Study data were part of the Oxford meta-analysis [14]. Patients were randomized in a 2:1 ratio between 1-year tamoxifen (30 mg per day) and no adjuvant therapy. Eligible patients were postmenopausal, <76 years of age, and had a T1–4, N0–3, M0 breast tumor [15] with no mastitis or palpable supra- or infraclavicular lymph nodes. After 1 year, for patients in the tamoxifen arm who were on study, a second randomization was performed to receive another 2 years of tamoxifen or to stop further treatment. From 1989, based on two interim analyses showing a significant improvement in recurrence-free survival among lymph node-positive patients, these node-positive patients were all allocated to the tamoxifen treatment arm (i.e., skipped the first randomization). In total, 1,662 patients were included. The patient characteristics and clinical outcome of tamoxifen treatment have been presented elsewhere [13].
We have traced tissue blocks of participating patients and recollected sufficient tumor material of 739 patients who did not differ in prognostic factors from the total group (Table S1). After revision of estrogen receptor α (ERα) status as assessed with immunohistochemistry (IHC), a total of 563 ERα-positive tumors were used for subsequent analysis. The number of patients in each treatment arm of randomization 1 and randomization 2, pre- and post-interim analysis, is shown in Figure S1.
Immunohistochemistry
Tissue microarrays (TMAs) were constructed using formalin-fixed paraffin-embedded (FFPE) tumor blocks. The TMAs were stained for ERα, progesterone receptor (PgR), and HER2. ERα and PgR were considered positive when ≥10 % of invasive cells showed nuclear reactivity. This cutoff was chosen because it is common practice in the Netherlands and in addition this would avoid the potential inclusion of basal-like tumors [16] in our analysis. HER2 was considered positive when membranous staining was score 3. In case of a membranous score of 2, chromogenic in situ hybridization (CISH) was performed on whole-tissue slides. For tumors that did not have sufficient cores in the TMA, whole slides were cut and could adequately be assessed for ERα (N = 60), PgR (N = 55), and HER2 (N = 36). Tumor grade was scored on a hematoxylin–eosin (HE)-stained slide using the modified Bloom–Richardson score [17].
DNA isolation
From paraffin-embedded tissue blocks, 10-μM-thick sections were cut and attached to microscope slides. A total of 10 slides per tumor were used for DNA isolation. Slides were deparaffinized in xylene, rehydrated, and stained with hematoxylin. The slides were incubated with sodiumthiocyanate overnight. Exact tumor location was circled by the pathologist on a HE-stained slide, which was used as a template. After adding a drop of tissue lysis buffer, tumor tissue was scraped from the slides, added to a 1.5-μl micro-centrifuge tube containing a 200-μl mix of tissue lysis buffer/proteinase K. This tube was incubated in a Thermomixer at 55 °C for 48 h. An additional 27-μl proteinase K (2 mg/μl) was added after 24 and 36 h. After 48 h, the tube was incubated at 80 °C for 10 min to inactivate proteinase K. After centrifuging, the supernatant was pipetted into a new tube. A total of 150 μl was purified using a QIAquick PCR purification kit. DNA isolation was successful for 535 tumor samples.
Genotyping
Genotyping for CYP2C19*2 (681G>A, rs4244285) and CYP2C19*17 (−806C>T, rs12248560) was performed on tumor DNA. The concordance between selected genotyping from FFPE-derived tumor DNA and DNA from serum has previously been established [18]. Taqman allelic discrimination assays were used (Applied Biosystems, Nieuwerkerk ad IJssel, The Netherlands) on an ABI Prism 7500 Sequence Detection system (Applied Biosystems). Each assay consisted of two allele-specific minor groove-binding (MGB) probes, labeled with fluorescent dyes VIC and FAM. The assay IDs are C_25986767_70 (CYP2C19*2) and C_469857_10 (CYP2C19*17). Thermal profile for genotyping consisted of 95 °C for 10 min, followed by 50 cycles of 15 s at 92 °C and 90 s at 60 °C. Genotypes were scored by allele-specific fluorescence using 7500 fast system SDS software (Applied Biosystems).
Statistics
Recurrence-free interval (RFI) was taken as the time from the date of (first) randomization until the occurrence of a local, regional, or distant recurrence or breast cancer-specific death [19]. Since these CYP2C19 variants are associated with breast cancer risk and the duration of treatment in this trial was relatively short to prevent the occurrence of a secondary breast cancer, patients with a secondary contra-lateral breast cancer were censored at the date of this occurrence (Table S2). In the subset of 563 ERα-positive patients, median follow-up of patients without a recurrence event is 7.8 years. Genotypes were tested for Hardy–Weinberg equilibrium using a Chi square test. The distribution of clinico-pathological characteristics by the CYP2C19*2 and CYP2C19*17 variants was evaluated using Chi square tests. Survival curves were constructed using the Kaplan–Meier method and compared using log-rank tests. To determine whether the benefit from tamoxifen was different in CYP2C19 (one or two *2 alleles vs. no *2 allele and one or two CYP2C19*17 alleles vs. no CYP2C19*17 allele) genotypes, covariate adjusted Cox proportional hazard regression models were constructed with an interaction between the treatment and the genotype. Treatment groups were defined according to the results of the first randomization (1–3 years of tamoxifen versus no adjuvant systemic treatment). The change in randomization that occurred after the interim analysis resulted in an enrichment of lymph node-positive patients in the group of tamoxifen-treated patients. Therefore, Cox proportional hazard regression models were stratified for nodal status. The following factors were included as covariates: age (≥65 vs. <65), grade (grade 3 vs. grade 1–2), tumor size (T3–T4 vs. T1–T2), HER2 status (positive versus negative), estrogen receptor expression (10–99 vs. 100 %), and progesterone status (positive versus negative). No adjustments for multiple testing were performed. To assess the prognostic value of the CYP2C19 genotypes, we performed covariate adjusted Cox proportional hazard regression (including lymph node status) in the subgroup of patients who were randomized to the control arm. This study complied with reporting recommendations for the tumor marker prognostic studies (REMARK) criteria [20] outlined in Table S3.
Results
Study population and genotypes
Adequate genotype data for CYP2C19*2 and CYP2C19*17 were available for 494 and 504 patients, respectively (Fig. S2). For CYP2C19*2, a total of 12 (2.4 %) patients were homozygous carrier and 127 (25.7 %) heterozygous carriers were found. The homozygote CYP2C19*17/*17 genotype was seen in 28 (5.6 %) of the patients, while 151 (30.0 %) were heterozygous. Genotype frequencies of CYP2C19*2 and CYP2C19*17 were in Hardy–Weinberg equilibrium (p = 0.86 and p = 0.06, respectively). A weak association between CYP2C19*2 and low ERα expression was observed (Table 1). A higher frequency of grade III was observed in patients carrying a CYP2C19*17 allele. (Table S4). In general, known prognostic factors were equally divided over the treatment arms for all genotypes (Tables S5, 6), with the exception of lymph node status which can be explained by the change in randomization. In patients without a CYP2C19*17 allele, the tamoxifen arm included more HER2-positive patients than the control arm, while in patients with a CYP2C19*17 allele, the patients in the control arm were younger than the patients in the tamoxifen arm.
Association of genotypes with tamoxifen response in ERα-positive patients
When stratified by nodal status, the tamoxifen effect in all 563 ERα-positive patients is 0.54 (95 % CI 0.36–0.83, p = 0.004). In univariate analysis, positive lymph node status was associated with an unfavorable outcome (HR 2.61, p < 0.001). No significant interaction between lymph node status and tamoxifen was observed.
We did not find a significant interaction between the CYP2C19*17 genotype and treatment (adjusted p for interaction = 0.62) (Table S7). However, a significant interaction between CYP2C19*2 genotype and treatment was found (both unadjusted and adjusted p for interaction = 0.04). Patients carrying one or two CYP2C19*2 alleles derived more benefit from tamoxifen than patients with no CYP2C19*2 allele (CYP2C19*2 carriers: adjusted HR = 0.26, p = 0.001; non-CYP2C19*2 carriers: adjusted HR = 0.64, p = 0.18; Figs. 1 and S3; Table 2). Performing a sensitivity analysis, using a 3-level factor for T-stage (T1, T2, and T3–4), did not substantially change these results (Table S8).
Kaplan–Meier survival analysis for recurrence-free interval according to tamoxifen treatment in patients without a CYP2C19*2 allele (a) and patients with a CYP2C19*2 allele (b). Hazard ratios, 95 % confidence intervals, and p values were based on multivariate Cox regression analysis, stratified for nodal status
Tamoxifen-untreated patients with a CYP2C19*2 variant allele (N = 33) had an unfavorable RFI when compared with tamoxifen-untreated patients without a CYP2C19*2 allele (N = 86). After correcting for the prognostic factors as described in the methods section, CYP2C19*2 remained an independent adverse prognostic factor (HR = 2.77, p = 0.01) (Table S9). We did not find a difference in RFI in control patients with a CYP2C19*17 allele (p = 0.91) (Table S10).
Conclusion and discussion
In this study population, postmenopausal breast cancer patients with a CYP2C19*2 variant allele had a worse prognosis, but derived more benefit from adjuvant tamoxifen treatment compared to patients without a CYP2C19*2 allele.
Previously, others have found discordant effects of CYP2C19*2 with respect to outcome after tamoxifen treatment. Okishiro et al. [21] found a favorable recurrence-free survival for Japanese patients treated with adjuvant tamoxifen who were homozygous for CYP2C19*2 or CYP2C19*3 variants, compared with patients with heterozygote or wild-type phenotype, although this difference was not significant (HR = 0.37, p = 0.21). The number of patients in that study was relatively small (N = 173) and the comparison that was made was different from our analysis, since we grouped homozygous and heterozygous CYP2C19*2 carriers together. In a study from van Schaik et al. [10], the CYP2C19*2 variant was assessed in three independent series of patients who were treated with tamoxifen for metastatic breast cancer. An increased time to treatment failure was observed for patients with the CYP2C19*2 variant. Ruiter et al. [22] analyzed CYP2C19*2 in a Dutch population-based cohort study. In this study, a subset of 80 female breast cancer patients who were treated with tamoxifen (adjuvant or palliative), CY2C19*2 carriers, had a better survival (HR = 0.26, p = 0.03). However, in a series of patients who were all treated with adjuvant tamoxifen, Schroth et al. [11] observed an unfavorable outcome for patients carrying a CY2C19*1, *2, or *3 allele compared to patients with a CYP2C19*17 allele. Since none of the above-mentioned studies included a (matched) control group, the predictive value of the polymorphism cannot be discerned from its prognostic value. In the adjuvant setting with recurrence-free survival as the endpoint, the prognosis is not only affected by the molecular subtype of the primary tumor but also by the occurrence of secondary primary breast cancer. Since CYP2C19*2 and CYP2C19*17 have been associated with, respectively, increased and decreased breast cancer risk [8], this might also affect the occurrence of secondary breast tumors, particularly in the case of relatively long follow-up. In addition, CYP2C19 variants might influence breast cancer prognosis by their lifelong affect on estrogen levels, which might potentially influence breast cancer molecular subtype [9].
We observed in our series that the CYP2C19*2 variant was associated with unfavorable prognosis. In the series of Ruiter et al., CYP2C19*2 was not significantly associated with breast cancer mortality in patients not using tamoxifen. We exclusively analyzed ERα-positive patients which may explain the inconsistency in our findings and those of Ruiter et al. To our knowledge, there are no other studies that analyzed the effect of this CYP2C19 variant on breast cancer prognosis.
Several explanations for the increased benefit from tamoxifen in patients who carry a CYP2C19*2 allele can be considered. Since the presence of this non-functional allele would expectedly result in a decrease in active metabolites and a reduced tamoxifen benefit, simply the variation in active tamoxifen metabolite levels is not an obvious explanation. Previously, it had been suggested that reduced isomerization of trans-endoxifen into the less potent cis-endoxifen in patients with a CYP2C19*2 allele may result in an increased response to tamoxifen therapy [10]. Since we did not observe an opposite association with tamoxifen response for patients carrying the highly active CYP2C19*17 variant, this is not likely the case. Moreover, CYP2C19 polymorphisms do not seem to significantly affect tamoxifen metabolite levels in breast cancer patients [23], which also argues against an effect mediated by tamoxifen metabolism.
It is more likely that women carrying a CYP2C19*2 genotype are susceptible to tumors that are highly dependent on estrogen signaling due to their lifelong higher exposure to estrogens [7] and would therefore be more sensitive to estrogen-inhibiting therapy. Since we did not have serum available from patients who participated in this trial, we were not able to obtain supportive data for this hypothesis in our series. We also did not have gene expression data available to test whether patients with a CYP2C19*2 variant had a different molecular subtype.
We did not correct for multiple testing. In early stage research, the concern is both to avoid type II as well as type I errors. It would be interesting to confirm our observations and further explore our hypothesis in randomized series of patients from whom genotype, gene expression, and serum are available, treated with and without anti-estrogens.
Another limitation of our study is the relatively short duration of tamoxifen therapy (1–3 years) in this retrospectively analyzed randomized trial. Currently, a minimal duration of 5 years is common clinical practice; however, we anticipate that the relative effects of the genotypes analyzed in this study will be similar for shorter and longer durations of endocrine therapy. The patients in our study randomized to adjuvant treatment received tamoxifen only (and no aromatase inhibitors), while currently most ERα-positive, postmenopausal breast cancer patients receive an aromatase inhibitor preceding or following tamoxifen treatment. Since we hypothesize that tumors from women with a CYP2C19*2 genotype are highly dependent on estrogen signaling, we expect an increased benefit from aromatase inhibitors as well in these patients. It would be interesting to analyze this question in randomized trials comparing aromatase inhibition with tamoxifen. The patients in our trial had not received adjuvant chemotherapy, and thereby the observed effect of the CYP2C19 genotype was not biased by adjuvant chemotherapy that might have cured endocrine-resistant patients.
In summary, CYP2C19 is primarily involved in endogenous steroid metabolism and may therefore influence tumorigenesis and sensitivity to endocrine therapy which has not yet been appreciated. We show that the presence of a CYP2C19*2 variant allele identifies those ERα-positive postmenopausal breast cancer patients who have an adverse prognosis, but will derive substantial benefit from adjuvant tamoxifen.
References
Lee AJ, Cai MX, Thomas PE, Conney AH, Zhu BT (2003) Characterization of the oxidative metabolites of 17beta-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology 144(8):3382–3398
Desta Z, Ward BA, Soukhova NV, Flockhart DA (2004) Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther 310(3):1062–1075
Yamazaki H, Shimada T (1997) Progesterone and testosterone hydroxylation by cytochromes P450 2C19, 2C9, and 3A4 in human liver microsomes. Arch Biochem Biophys 346(1):161–169
Farhat GN, Cummings SR, Chlebowski RT, Parimi N, Cauley JA et al (2011) Sex hormone levels and risks of estrogen receptor-negative and estrogen receptor-positive breast cancers. J Natl Cancer Inst 103(7):562–570
Tamminga WJ, Wemer J, Oosterhuis B, de Zeeuw RA, de Leij LF et al (2001) The prevalence of CYP2D6 and CYP2C19 genotypes in a population of healthy Dutch volunteers. Eur J Clin Pharmacol 57(10):717–722
Li-Wan-Po A, Girard T, Farndon P, Cooley C, Lithgow J (2010) Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br J Clin Pharmacol 69(3):222–230
Gjerde J, Geisler J, Lundgren S, Ekse D, Varhaug JE et al (2010) Associations between tamoxifen, estrogens, and FSH serum levels during steady state tamoxifen treatment of postmenopausal women with breast cancer. BMC Cancer 10:313
Justenhoven C, Hamann U, Pierl CB, Baisch C, Harth V et al (2009) CYP2C19*17 is associated with decreased breast cancer risk. Breast Cancer Res Treat 115(2):391–396
Dunbier AK, Anderson H, Ghazoui Z, Folkerd EJ, A’Hern R et al (2010) Relationship between plasma estradiol levels and estrogen-responsive gene expression in estrogen receptor-positive breast cancer in postmenopausal women. J Clin Oncol 28(7):1161–1167
van Schaik RH, Kok M, Sweep FC, van Vliet M, van Fessem M et al (2011) The CYP2C19*2 genotype predicts tamoxifen treatment outcome in advanced breast cancer patients. Pharmacogenomics 12(8):1137–1146
Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T et al (2007) Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol 25(33):5187–5193
Michalides R, van Tinteren H, Balkenende A, Vermorken JB, Benraadt J et al (2002) Cyclin A is a prognostic indicator in early stage breast cancer with and without tamoxifen treatment. Br J Cancer 86(3):402–408
Vermorken JB, Burgers JMV, Taat CW, van de Slee PHT, Hennipman A et al (1998) Adjuvant tamoxifen in breast cancer: interim results of a comprehensive cancer center Amsterdam trial. Breast Cancer Res Treat 50(3):223
Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365(9472):1687–1717
World Health Organisation (2011) World Health Organization Histological Typing of Breast Tumours 1981, 2nd edn. World Health Organisation, Geneva
Iwamoto T, Booser D, Valero V, Murray JL, Koenig K et al (2012) Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1 to 10 % ER-positive by immunohistochemistry. J Clin Oncol 30(7):729–734
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19(5):403–410
Sjoholm MI, Hoffmann G, Lindgren S, Dillner J, Carlson J (2005) Comparison of archival plasma and formalin-fixed paraffin-embedded tissue for genotyping in hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev 14(1):251–255
Hudis CA, Barlow WE, Costantino JP, Gray RJ, Pritchard KI et al (2007) Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol 25(15):2127–2132
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M et al (2005) Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 97(16):1180–1184
Okishiro M, Taguchi T, Jin KS, Shimazu K, Tamaki Y et al (2009) Genetic polymorphisms of CYP2D6 10 and CYP2C19 2, 3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer 115(5):952–961
Ruiter R, Bijl MJ, van Schaik RH, Berns EM, Hofman A et al (2010) CYP2C19*2 polymorphism is associated with increased survival in breast cancer patients using tamoxifen. Pharmacogenomics 11(10):1367–1375
Lim JS, Chen XA, Singh O, Yap YS, Ng RC et al (2011) Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br J Clin Pharmacol 71(5):737–750
Acknowledgments
We would like to thank Judy Jacobse for her help with DNA isolation procedures. We thank all pathology departments throughout the Netherlands for submission of FFPE tumor blocks. This study was sponsored by TI Pharma (Project Number T3-502) and by A Sister’s Hope.
Disclosures
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Beelen, K., Opdam, M., Severson, T.M. et al. CYP2C19*2 predicts substantial tamoxifen benefit in postmenopausal breast cancer patients randomized between adjuvant tamoxifen and no systemic treatment . Breast Cancer Res Treat 139, 649–655 (2013). https://doi.org/10.1007/s10549-013-2568-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-013-2568-0