Abstract
An integrated microfluidic system has been developed for rapid enumeration of CD4 + T lymphocytes at point-of-care (POC) settings. A concise microfluidic chip, which consists of three separate chambers, respectively, for reaction, detection and waste storage, is developed to automate CD4 detection. To simplify CD4 + T lymphocyte enumeration, a single polycarbonate bead immobilized with CD4 antibody is adopted by the microfluidic chip to capture the CD4 antigen in the lysed testing sample. Desired performance is achieved by actuating the single bead for efficient mixing, as well as transferring it between different reaction chambers to reduce non-specific reaction. A controllable external magnetic field is applied to drive the single bead with a built-in ferrous core for different purposes. Chemiluminescence reaction is implemented in an independent chamber to reduce non-specific binding of enzyme. A simple flow control strategy is adopted to conveniently release the waste reagent into the waste storage chamber by just opening the vent hole without actively pumping. A sensitive CCD camera is used to collect the reaction signal by taking picture from the single bead, and then the signal intensity is further analyzed for CD4 + T lymphocyte enumeration. Experimental results show that rapid, convenient, accurate and low-cost CD4 + T lymphocyte enumeration can be obtained with the developed microfluidic system at POC test.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
By 2015, it is estimated that more than 36.7 million people in the world have been infected by human immunodeficiency virus (HIV) (Charlson et al. 2016). For HIV infected hosts without any treatment, their CD4 levels will gradually decreases and eventually they will be identified as patients with Acquired Immune Deficiency Syndrome (AIDS). Therefore, the level of CD4 + T lymphocytes provides valuable clinical information for the diagnosis and therapy of HIV/AIDS. As suggested by WHO (Word Healthy Organization), HIV infected hosts should receive antiretroviral therapy (ART) when their CD4 levels are below a threshold value, for example, 350 cells/μL to prevent serious complications caused by AIDS (Aina et al. 2005). For the good living quality, it is suggested that the CD4 level of HIV infected hosts should be monitored four times per year (Boyle et al. 2012). As a critical tool for HIV/AIDS diagnosis and therapy, CD4 + T lymphocyte enumeration has great significance in HIV monitoring, assessment of opportunistic infection, and efficacy evaluation of antiretroviral treatment (Douek et al. 2002).
As a gold standard, flow cytometry is the first choice for accurate enumeration of CD4 + T lymphocytes. However, flow cytometers are normally expensive, bulky and complicated, which significantly limit their applications (Piyasena and Graves 2014). Well trained personnel are normally required to operate flow cytometers and analyze the experimental data. Because of its dependence on infrastructural facilities, such as electricity, refrigerator and deionized water, CD4 + T lymphocyte enumeration with flow cytometers has to be performed in central labs. In contrast, manual CD4 + T lymphocyte enumeration can be completed with a microscope and hemocytometer (Carella et al. 1995). Because of the trivial procedure, manual CD4 counting could result in low efficiency and high counting error even though the cost is relatively low. ‘Lab-on-a-chip’ systems are able to integrate and automate the bio-analytical process with a microfluidic network (Sposito et al. 2016; Kokkinis et al. 2017). Ideally, integrated and easy-to-operate microfluidic systems enable the complicated diagnostic procedure to be performed in an automatic way by less-trained personnel at low resource settings in POCT (Kyungsup et al. 2016; Mohammad et al. 2017; Kunstmann-Olsen et al. 2016). To achieve rapid, convenient and low-cost CD4 + T lymphocyte enumeration in POCT, different methods for CD4 counting with microfluidic chips have been intensively studied (Du et al. 2016; Beck et al. 2012; Masahito et al. 2012).
One way is to first isolate and detect each single CD4 cell based on its physical, optical or electrical properties in microfluidic chips, and then CD4 + T lymphocyte enumeration is performed by recognizing each CD4 lymphocyte one by one (Yun et al. 2010; Zhu et al. 2013). For example, first CD4 cells are specifically captured by antibodies immobilized on the surface of the reaction chamber, and then image-based CD4 + T lymphocyte enumeration can be performed by identifying each cell through specific fluorescence labeling (Ymeti et al. 2007; Li et al. 2010). In an alternative way, CD4 cells in the test sample can be separated in a microfluidic channel with properly designed dimensions to guide them to go through the specific detection zone one by one, and CD4 + T lymphocyte enumeration can be achieved by recognizing each cell based on its own fluorescence or impedance signal (Watkins et al. 2011; Wu et al. 2008). Normally, to perform CD4 + T lymphocyte enumeration based on differentiation to single cell, elaborative and complicated optical or electrical module has to be developed to ensure the reasonable recognition resolution and the high signal-to-noise ratio (Moon et al. 2009; Ozcan and Demirci 2008).
In contrast, the other way is to perform CD4 + T lymphocyte enumeration based on the integral physical or chemical properties of the entire group of target cells instead of recognizing each CD4 cell in microfluidic chips (Kiesel et al. 2011; Gohring and Fan 2010). For example, first CD4 cells are separated from other types of cells in a reaction chamber, and then CD4 + T lymphocyte enumeration is performed by chemically lysing CD4 cells and measuring the impedance of the solution with lysed cells (Cheng et al. 2007a, b). Alternatively, CD4 + T lymphocyte enumeration can be implemented by measuring the change of electrochemical impedance between different electrodes where CD4 cells are specifically captured by immobilized antibodies (Jiang and Spencer 2010). In a more straightforward way, CD4 + T lymphocyte enumeration can be performed by packaging CD4 cells captured by magnetic beads into a specific space with magnetic force or gravity, and then the number of CD4 cells can be figured out through the physical dimension, e.g., length or height of the densely accumulated cells (Glynn et al. 2014; Zachariah et al. 2011). Using chemiluminescence as a detection method, semiquantitative CD4 + T lymphocyte enumeration can be performed when CD4 cells are captured by densely arranged microfabricated traps coated with anti-CD4 antibodies in a microfluidic reactor (Wang et al. 2009). In principle, CD4 + T lymphocyte enumeration based on the integral physical or chemical properties of the entire group of CD4 cells can potentially reduce the complexity and cost of the microfluidic diagnosis system, which is especially beneficial to POCT (Cheng et al. 2007a, b; Willyard 2007).
In this paper, we reported a rapid, simple, convenient and easy-to-use CD4 + T lymphocyte enumeration method based on a microfluidic system with a single bead in POCT. Different cells in the blood sample including red cells, CD4 and other cells are first lysed with lysis buffer, and then CD4 antigen in the lysed sample is specifically captured by a single bead with a high surface-to-volume ratio to ensure the detection sensitivity. An easy-to-operate microfluidic system with three separate chambers is developed to facilitate CD4 + T lymphocyte enumeration without the need of active pumping. The single bead with a built-in ferrous core can be easily actuated by a controllable external magnetic field in the microfluidic chip, which facilitates not only mixing with sample or reagent, but also transport between the reaction and the detection chambers. Chemiluminescence assay is adopted for CD4 + T lymphocyte enumeration because of low background with the detection signal. CD4 + T lymphocyte enumeration with the developed microfluidic system is achieved by analyzing the chemiluminescence signal intensity of the single bead collected by a CCD camera, and its performance is validated with different testing samples.
2 Material and methods
2.1 Outline of the POC CD4 + T lymphocyte enumeration system
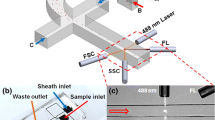
As shown in Fig. 1, an integrated microfluidic system is developed to perform CD4 + T lymphocyte enumeration in a simple, rapid, convenient and low-cost way at point-of-care testing. As shown in Fig. 1a, for CD4 + T lymphocyte enumeration, CD4 antigen from the lysed blood sample is detected by a single-bead of 2 mm in diameter in the format of sandwiched chemiluminescence immunoassay. CD4 antibody is immobilized on the large surface of the polycarbonate bead for CD4 antigen capturing. Horseradish peroxidase (HRP) enzyme conjugated with the second CD4 antibody is used for chemiluminescence analysis when it is specifically captured by the CD4 antigen. An iron ball with a diameter of 0.6 mm is embedded in the single bead as the ferrous core in order to flexibly actuate the single bead for different purposes. As shown in Fig. 1b, based on a concise microfluidic chip, a simple actuation method with magnetic control is developed not only to move the single bead up and down for mixing enhancement, but also to transport the single bead between different reaction chambers for reduced non-specific reaction. Meanwhile, as shown in Fig. 1b, with CCD camera based detection, a quantification method for CD4 + T lymphocyte enumeration based on chemiluminescence image collection and analysis to the single bead with captured CD4 antigen is developed.
2.2 Microfluidic chip with a single bead for CD4 + T lymphocyte enumeration
An integrated microfluidic chip is developed to achieve rapid CD4 + T lymphocyte enumeration based on the single bead. The real image of single beads is as shown in Fig. 2a. As shown in Fig. 2b, the microfluidic chip includes a reaction module (41 × 35 × 8 mm) made by black PMMA and a waste storage module (50 × 30 × 16 mm) made by transparent PMMA, and both of them are fabricated by acetonitrile based solvent bonding after laser ablation. As shown in Fig. 2b, the reaction module consists of a reaction chamber (20 × 5 × 4 mm) for the sandwiched immunoassay, a detection chamber (15 × 5 × 4 mm) for chemiluminescence analysis, and a release channel to discharge the waste reagent into the waste storage chamber located in the waste storage module. Lysate sample or reagent is loaded sequentially into the reaction chamber to react with the functionalized single bead for CD4 + T lymphocyte enumeration. As shown in Fig. 2c, for the magnetic field control, a permanent magnet, which is mounted on a linear moving stage, is used to move vertically the single bead up and down for mixing when it is incubated with the lysate sample, HRP enzyme or the wash buffer to achieve sensitive detection in a short time. As shown in Fig. 2d, there has a connection hole between the reaction chamber and the waste storage module to release the waste reagent if necessary. As shown in Fig. 2e-f, after reaction with HRP enzyme, the washed single bead is first lifted up along the sloping wall of the reaction chamber and then transported into the detection chamber via the magnetic control when the magnet first goes up and then goes down. Once the single bead with the gravity (F2) reaches the top of the side wall, it is drawn to the other chamber because of the magnetic force (F3) when the force from the sloping wall (F1) disappears. As shown in Fig. 2g, the detection chamber is filled with mixed substrate buffers to react with the HRP enzyme captured on the surface of the single bead for chemiluminescence when the single bead goes up and down for enhanced mixing with magnetic actuation.
Microfluidic chip with a single bead for CD4 + T lymphocyte enumeration. a The illustration of the chemiluminescence analysis with the single bead. The bead is immobilized with CD4 antibody first, then the CD4 antibody reacts with the surrounding CD4 antigen in the lysate sample. Finally, HRP enzyme is specifically captured by the single bead through the second CD4 antibody before it speeds up the chemiluminescence with the mixed substrate buffers. b The detailed structure of the reaction and the waste storage modules. c-d Sample, HRP incubation or wash step with mixing, and the waste reagent discharging. e-g The single bead transport and the subsequent chemiluminescence incubation in the detection chamber with mixing
As shown in Fig. 2b, with the reaction module consisting of two independent chambers, the non-specific reaction with the HRP enzyme remained on the wall of the reaction chamber is effectively avoided when the single bead is transported from the reaction chamber into the detection chamber, which is helpful to achieve reasonable detection with low background. As shown in Fig. 2b, a simple flow control strategy is adopted to release the waste reagent with its own gravity by opening the vent hole located on the waste storage module with a motorized linear actuation part. Once the vent hole is opened, the air pressure is balanced, and the waste reagent is released to the waste storage chamber through the drainage channel due to the gravity. If the vent hole is inversely closed, reagent will be kept in the reaction chamber.
2.3 Portable instrument for CD4 + T lymphocyte enumeration with microfluidic chip
As shown in Fig. 3, a portable instrument is developed to perform CD4 + T lymphocyte enumeration with the developed microfluidic chip. As shown in Fig. 3a, only two linear actuation modules, which are respectively actuated by two linear DC motors, are required to perform the whole operation for CD4 + T lymphocyte enumeration. The vertical linear motor is responsible for moving vertically the single bead up and down for enhanced mixing inside the reaction or the detection chamber through a combined permanent magnet from the outside wall of the reaction module. Meanwhile, the vertical linear motor is also used to transport the single bead from the reaction chamber to the detection chamber with the same permanent magnet. In contrast, the horizontal linear motor is responsive for opening or closing the vent hole located in the waste storage module through a combined rubber pin for the purpose of the flow control. Once the vent hole is sealed from outside by the rubber pin when it is actuated by the horizontal linear motor, the reagent will stay inside the reaction chamber. Otherwise, the waste reagent will be released into the waste storage chamber because of the gravity once the vent hole is unsealed when the rubber pin is retracted by the horizontal linear motor. With a simplified flow control strategy, operation for CD4 + T lymphocyte enumeration with the developed microfluidic chip can be conveniently performed with two linear motors without complicated fluid pumping and valving.
Instrument for CD4 + T lymphocyte enumeration with microfluidic chip. a Schematic of the actuation part with two linear motor modules. b Schematic of the chemiluminescence signal collection with a CCD camera. c Picture of the integrated instrument system. d Custom software interface for CCD camera control
As shown in Fig. 3b, a 16-bit Charge Coupled Device (CCD) camera (ST-1603, SBIG), which is located above the detection chamber, is used to take the chemiluminescence picture from the single bead inside the microfluidic chip. Custom software developed with Visual Studio 2010 is used to control the CCD camera with a laptop. As shown in Fig. 3d, an operation interface is provided to set various parameters for image based detection, for example, exposure time, sampling cycle, picture storage, signal intensity analysis and display. A custom image processing algorithm was developed to analyze the chemiluminescence signal intensity of the single bead automatically.
As shown in Fig. 3c, a portable integrated instrument consisting of an operation module, a CCD camera and a smartphone, was developed to perform CD4 + T lymphocyte enumeration with the microfluidic chip. Alternatively, the smartphone camera has also been used to detect the chemiluminescence signal, but it was not successful because of both the low amplitude of the chemiluminescence signal and the low sensitivity of the smartphone camera. The operation module including mechanical parts, electrical parts and other accessory parts is enclosed in a small box to avoid negative effect from the environmental light. Especially, a microcontroller system was developed to control the two linear motors to perform the whole operation for CD4 + T lymphocyte enumeration. Custom application software running on a smartphone (B880, ZTE Corporation) is developed with JAVA to provide the operation interface for CD4 + T lymphocyte enumeration. To allow the smartphone to guide the microcontroller system to implement each operation step, Bluetooth communication is established between two of them.
2.4 Sandwiched chemiluminescence immunoassay in microfluidic chip
First, to detect the CD4 antigen for CD4 + T lymphocyte enumeration, the blood sample (30 μL) is lysed with lysis buffer (210 μL), which is also helpful to avoid interference from a large quantity of red blood cells when the whole blood sample is used. Second, 240 μL lysate sample is manually added into the reaction chamber with a single bead immobilized with CD4 antibodies and both of them are incubated for around 15 min. Meanwhile, the single bead is moved up and down continuously with a speed of 1.3 mm/s through the magnetic control for specific binding of CD4 antigen from the lysate sample with enhanced mixing. After that, the waste reagent is released into the waste storage chamber by opening the vent hole. In the following steps, the single bead is always moved up and down (with a speed of 1.3 mm/s) continuously through the magnetic control when it is incubated with other reagent, for example the wash buffer, HRP enzyme or the mixed substrate buffers. And then, the single bead is washed with 260 μL wash buffer in one minute, and the wash step is repeated for another four times. For each wash, the wash buffer is added into the reaction chamber manually, and then it is released into the waste storage chamber after incubation by opening the vent hole. Third, 160 μL HRP enzyme is manually added into the reaction chamber, and the single bead is incubated with HRP conjugated the second CD4 antibody for specific enzyme binding for around 15 min. After that, the waste reagent is released into the waste storage chamber by opening the vent hole. And then, the single bead is washed with 260 μL wash buffer in one minute, and the wash step is repeated for another four times. Finally, after the single bead transport, 100 μL of two mixed substrate buffers with 50 μL for each of them are manually added into the detection chamber with the single bead, and the chemiluminescence with the mixed substrate buffers around the single bead is significantly speeded up by the HRP enzyme captured on the surface of the single bead because of catalysis effect.
Therefore, CD4 + T lymphocyte enumeration can be achieved by quantitatively analyzing the intensity of the collected chemiluminescence signal from the single bead. For CD4 + T lymphocyte enumeration with the developed microfluidic chip, the incubation time is significantly reduced because of the introduced mixing with the outside magnetic control to the single bead. With the optimized sandwiched chemiluminescence immunoassay in the developed microfluidic chip, CD4 + T lymphocyte enumeration can be completed within just 45 min even including collection and analysis of the chemiluminescence signal, which is beneficial to POCT.
2.5 Image processing of the single bead for CD4 + T lymphocyte enumeration
In principle, the concentration of the CD4 antigen is directly related to the chemiluminescence signal intensity on the single bead. Therefore, it is important to accurately analyze the chemiluminescence signal on the single bead for quantitative detection. A custom image processing algorithm was developed to analyze the signal intensity. As shown in Fig. 4, the single bead in the chemiluminescence image was identified first, and then its mean gray value subtracted by the background was regarded as the detection result.
As shown in Fig. 4a, the area with the single bead from the original chemiluminescence image with the size of 100 × 100 pixels is processed for quantitative analysis. First, the top boundary of the single bead is identified. As shown in Fig. 4b and c, the distribution of the gray value of the single bead and its surrounding background is analyzed when a horizontal sampling line moves down with a step of one pixel from the original position (0, 0). The top boundary of the single bead can be identified when the gray value distribution on one specific sampling line satisfies with a predefined condition.
The average gray value of all pixels on each horizontal sampling line, for example, y = y i , is defined as Aver,
where f(x, y i ) represents the gray value of the pixel at (x, y i )on the sampling line.
When the sampling line moves down, the top boundary of the single bead will be identified once the distribution of the gray value along a specific sampling line (y = y 0 ) satisfies with the following condition based on the difference of the gray value between the single bead and the surrounding background,
where a with an empirical value of 2 is a coefficient to improve the robustness, and (x0, y0) is an identified position on the recognized sampling line (y = y 0 ) defining the top boundary. Similarly, the bottom boundary of the single bead can be identified as the bottom boundary.
Next, the left and right boundaries of the single bead are identified based on the gradient of the gray value on all the sampling lines between the top and bottom boundaries of the single bead. Boundary searching starts from the sampling line defining the location of the top boundary, e.g., y = y 0 . Since x0 is the approximate left boundary of the sampling line, the empirical boundary searching region is limited between (x0−10, y0) and (x0 + 20, y0) based on the known size of the single bead. The gray value gradient between each two adjacent pixels within the searching region is defined as following,
The maximum (ΔKmax) and minimum (ΔKmin) values of ΔK(x) can be determined accordingly with the gray value gradient analysis, which respectively corresponds to the left (x L ) and right (x R ) boundaries of the sampling line. Similarly, the left and right boundaries on the rest sampling lines in the searching region can be identified. Finally, the single bead can be identified from the chemiluminescence image based on the determined boundaries of each sampling line in the searching region. The mean gray value of the single bead subtracted by the background is defined as the final detection result.
3 Results and discussion
For CD4 + T lymphocyte enumeration based on the sandwiched chemiluminescence immunoassay, a quantification model between the concentration of CD4 antigen in the lysate sample and the corresponding CD4 + T lymphocyte count in the blood sample is established with flow cytometry. It is found that 1 RU/μL (RU/μL is a relative unit calibrated by Beijing Wantai Biological Pharmacy Enterprise Co., Ltd.) of CD4 antigen concentration corresponds to 1 cell/μL of CD4 + T lymphocyte count in the test sample. Therefore, the number of CD4 + T lymphocytes in a unit volume can be figured out based on the concentration of CD4 antigen which can be achieved with developed microfluidic system.
3.1 Comparison of chips fabricated with different materials
Polymethyl methacrylate (PMMA) is a widely used polymer material in the field of microfluidic chip applications. Two types of PMMA, one transparent and the other with black color were compared with each other for optimal signal detection in chemiluminescence analysis. Two reacted single beads with high concentration test samples were respectively put into two independent detection chambers belonging to two different microfluidic chips, and a CCD camera was used to take picture from each of them for chemiluminescence signal collection. In the experiment, lysate of white blood cells were used as the test sample. As shown in Fig. 5, the detection result with the black material is much more desirable than that with the transparent material. Normally the transparent material is preferred in microfluidic chip fabrication because it is convenient for observation from outside. However, as shown in Fig. 5b, there has significant signal reflection from the inside wall of the detection chamber made from transparent PMMA, which contributes to an extraordinarily high and complicated background. Therefore, black PMMA with low and clear background around the single bead (as shown in Fig. 5a) was chosen because chemiluminescence analysis with a high signal-to-noise ratio can be achieved.
3.2 Dynamics analysis for Chemiluminescence based CD4 + T lymphocyte enumeration
In order to achieve accurate CD4 + T lymphocyte enumeration with chemiluminescence analysis, the dynamics of chemiluminescence reaction should be analyzed. Before that, two different operation modes respectively with diffusion based mixing and active mixing were compared between each other. For the natural diffusion based mixing, the single bead remained static in reaction, while for the active mixing, the single bead was moved up and down to introduce efficient mixing in reaction. As shown in Fig. 6, with active mixing, the signal amplitude subtracted by the background from the single bead is significantly higher than that achieved with the natural diffusion. The experiment was repeated for three times, and similar results were attained.
As shown in Fig. 6, when active mixing is introduced by moving the single bead up and down continually, the signal amplitude can be increased up to 1.73 times (from 2236 to 3865) comparing to the natural diffusion mode. Therefore, with active mixing, it will take short time for the chemiluminescence signal to reach a reasonably high reading, which could decrease the entire detection time for CD4 + T lymphocyte enumeration. In the following experiments, active mixing was always introduced for optimized performance.
Time response experiments with changing chemiluminescence signal intensity were performed to find out the proper time window for chemiluminescence detection with CCD camera. Two different samples respectively with two different CD4 antigen concentrations, 1200 RU/μL and 600 RU/μL, were tested. In the experiment, lysate of white blood cells were used as the test sample. Once the mixed substrate solution was added into the detection chamber for chemiluminescence, a CCD camera started to take picture from the single bead with an exposure time of 60 s continuously within seven min. Chemiluminescence signal intensities of the single bead subtracted by the background at different time were analyzed with the above mentioned intelligent image processing algorithm.
As shown in Fig. 7, for both single beads respectively with two different test samples, the chemiluminescence signal intensities have no significant change within the first two min, and then the signal starts to decrease with the elapse of time. As shown in Fig. 7, for the single bead with the high or low concentration test sample, the signal intensity drops from 6690 to 5215 gradually or from 3460 to 2923 gradually in the whole detection period. When the concentrations of the mixed substrate buffers drop down gradually because of chemiluminescence reaction, the signal intensity of chemiluminescence around the single bead will gradually decrease with the continuing HRP consumption. Therefore, to achieve sensitive CD4 + T lymphocyte enumeration with high stability and reliability, the mean value of the chemiluminescence signal intensities from the first two min are adopted as the detection result in all the tests.
3.3 Dose response curve of CD4 + T lymphocyte enumeration with the developed microfluidic chip system
Dose response curve of CD4 + T lymphocyte enumeration based on the developed microfluidic chip system is established with different test samples with low to high concentrations including 75 RU/μL, 150 RU/μL, 300 RU/μL, 600 RU/μL, and 1200 RU/μL. In the experiments, lysate of white blood cells were used as the test sample. Meanwhile, lysis buffer was used for negative control test. Experiments with each concentration were repeated for at least three times. The dose response curve of CD4 + T lymphocyte enumeration with the developed microfluidic system is shown in Fig. 8.
As shown in Fig. 8, the signal intensity of the single bead becomes stronger when the concentration of the test sample increases. For negative test, the signal intensity of the single bead is much close to the background, which is totally different from the positive test. For CD4 + T lymphocyte enumeration at POC settings, 75 RU/μL, e.g., 75 cells/μL, is adopted as the limit of quantitative detection with the developed microfluidic system. Chemiluminescence signal intensities of the single bead subtracted by the background for different test samples were analyzed with the above mentioned intelligent image processing algorithm. As shown in Fig. 8, with the dose response curve, a desired linear fitting model with exponential logarithm (R2 = 0.9983) between the CD4 antigen concentration or the CD4 + T lymphocyte count and the chemiluminescence signal intensity can be properly established. One test sample with known CD4 antigen concentration of 400 RU/μL was used to verify the accuracy of the linear fitting model, and the test result was 391 RU/μL with the detection error of 2.25%. Therefore, with the developed dose response curve, the single-bead based microfluidic system can perform accurate CD4 + T lymphocyte enumeration within the reasonable detection range between 75 RU/μL and 1200 RU/μL, or between 75 cells/μL and 1200 cells/μL in POCT.
For HIV-infected patients, as the clinically critical threshold, 350 cells/μL of CD4 count is regarded as an optimal condition to start ART. Meanwhile, 200 cells/μL of CD4 count is used as a clinical decision point for monitoring of CD4 cells in HIV-infected patients. The developed microfluidic chip system for CD4 + T lymphocyte enumeration is able to provide accurate monitoring of CD4 cells between 75 cells/μL and 1200 cells/μL, which is desired for effective monitoring and clinical diagnosis of HIV-infected patients.
3.4 CD4 + T lymphocyte enumeration with whole blood sample on microfluidic chip
To further evaluate the performance of the developed microfluidic system, totally seven whole blood samples (provided by Beijing Wantai Biological Pharmacy Enterprise Co., Ltd.) were tested. For each test, 30 μL whole blood sample was first mixed with 210 μL lysis buffer for 10 s, and then totally 240 μL lysate was added into the reaction chamber with a single bead. For comparison, for the seven whole blood samples, CD4 + T lymphocyte enumeration with flow cytometry were performed in parallel. The correlation curve between the developed method with microfluidic system and flow cytometry is shown in Fig. 9.
As shown in Fig. 9, with the developed microfluidic system, comparable detection results for multiple test samples to those of flow cytometry can be achieved. An acceptable correlation model (R2 = 0.9597) between the microfluidic system and flow cytometry is properly developed, and the mean error for all the test samples is around 9.5%. Since CD4 is also expressed on monocytes, a small percentage of CD4 antigen in the lysate sample will come from monocytes although most of them from CD4 + T lymphocytes. For most of existing microfluidic systems, monocytes are normally isolated from the CD4 + T lymphocyte before enumeration. To reduce the complexity of the developed microfluidic chip, instead of separating monocytes from CD4 + T lymphocytes first, both of them are lysed in the whole blood sample simultaneously. In principle, the undesired contribution of CD4 antigen from monocytes to the test result can be regarded as a systematic error in CD4 + T lymphocyte enumeration. Therefore, the error of CD4 + T lymphocyte enumeration caused by monocytes can be partly compensated by the dose response model based on the calibrated test samples. To perform CD4 + T lymphocyte enumeration in POCT, a compromise between the precision and the simplification has to be made for balanced performances.
As a proof of concept, it has been demonstrated that successful CD4 + T lymphocyte enumeration with the developed microfluidic system for whole blood test samples can be achieved with reasonable accuracy within 45 min. Comparing to the existing CD4 + T lymphocyte enumeration method with flow cytometry or complicated microfluidic chips, with the developed microfluidic chip and its portable companion instrument, simple, convenient, low-cost, sensitive and immediate CD4 + T lymphocyte enumeration can be achieved at POC settings.
4 Conclusions and outlook
An automatic CD4 + T lymphocyte enumeration method based on an integrated microfluidic system has been developed for rapid, simple, accurate, convenient and low-cost CD4 counting. An integrated microfluidic chip made from PMMA with two independent reaction chambers and one waste storage chamber is developed to perform CD4 + T lymphocyte enumeration with the assistance of a single bead of 2 mm in diameter. CD4 antigen in the lysed sample is captured by the single bead immobilized with CD4 antibody. Based on the configuration with double reaction chambers, after incubation with the test sample and HRP enzyme in turn in the first chamber, the single bead is transported into the second chamber, e.g., detection chamber for chemiluminescence reaction, which could significantly reduce non-specific reaction. To improve the detection sensitivity with reduced reaction time, instead of just relying on natural diffusion, the single bead with a built-in ferrous core is moved up and down in the reaction chamber by a controllable outside magnetic field to enhance the mixing between the single bead and the test sample or other reagent. Meanwhile, the outside controllable magnetic field is also used to transport the single bead between two separate reaction chambers. The waste reagent from each reaction step is released into the waste storage chamber with gravity by simply controlling the vent hole without any actively pumping.
A CCD camera is adopted to collect chemiluminescence signal by taking picture from the single bead with a properly set exposure time, and a custom image processing algorithm is developed to quantitatively analyze the detection signal for CD4 + T lymphocyte enumeration. It is found that the microfluidic chip has to be made from black material, for example black PMMA to avoid signal reflection in chemiluminescence image detection. The dynamics of chemiluminescence reaction reveals that the signal intensity of the single bead will gradually decrease with the elapse of time, which suggests that a proper time window for chemiluminescence signal collection should be chosen. Dose response curve shows that a satisfied quantification model is established with the developed microfluidic system for accurate CD4 + T lymphocyte enumeration. Experimental results with multiple whole blood test samples demonstrate that rapid and accurate CD4 + T lymphocyte enumeration can be achieved with the developed microfluidic system in a convenient way.
In summary, an easy-to-use and simple microfluidic system for CD4 + T lymphocyte enumeration including disposable microfluidic chips and a low-cost companion actuation module is successfully developed. Instead of relying on complicated and elaborate pumping, optical and electrical modules, CD4 + T lymphocyte enumeration is conveniently achieved with a single-bead based microfluidic chip which is simply actuated by two liner motors. Moreover, detection time for CD4 + T lymphocyte enumeration is significantly shortened to 45 min by the enhanced mixing between the single bead and the test sample or the reagent without sacrifice of the detection signal intensity. Therefore, rapid and low-cost CD4 + T lymphocyte enumeration can be achieved with the much simplified microfluidic system, which potentially could be beneficial to POC HIV/AIDS diagnosis and therapy in resource poor settings.
In the next step, a simpler signal detection strategy will be developed to replace the CCD camera with a high sensitive photodiode to further decrease the system cost and size. Meanwhile, different strategies for reagent storage on the microfluidic chip will be studied to develop a fully integrated microfluidic system for CD4 + T lymphocyte enumeration. In principle, by fabricating different beads, e.g. functionalization with different proteins for specific antigen or antibody capturing, the developed microfluidic system can be conveniently modified for other applications, for example, POC diagnosis of different diseases based on immunoassay.
References
O. Aina, J. Dadik, M. Charurat, P. Amangaman, S. Gurumdi, E. Mang, R. Guyit, N. Lar, P. Datong, C. Daniyam, P. Kanki, A. Abimiku, Clin. Diagn. Lab. Immunol. 12, 525 (2005)
M. Beck, S. Brockhuis, V.N. van der, C. Breukers, J. Greve, L.W. Terstappen, Lab Chip 12, 167 (2012)
D.S. Boyle, K.R. Hawkins, M.S. Steele, M. Singhal, X. Cheng, Curr. Trends Biotechnol. 30, 45 (2012)
A.V. Carella, M.W. Moss, V. Provost, T.C. Quinn, Clin. Diagn. Lab. Immunol. 2, 623 (1995)
F.J. Charlson, H.E. Erskine, A.J. Ferrari, J. Leung, H.A. Whiteford, A.A. Abajobir, L.D. Knibbs, R. Lalloo, J.G. Scott, Y. Guo, Lancet 388, 1545 (2016)
X. Cheng, Y.S. Liu, D. Irimia, U. Demirci, L. Yang, L. Zamir, W.R. Rodriguez, M. Toner, R. Bashir, Lab Chip 7, 746 (2007a)
X. Cheng, D. Irimia, M. Dixon, J.C. Ziperstein, U. Demirci, L. Zamir, R.G. Tompkins, M. Toner, W.R. Rodriguez, JAIDS 45, 257 (2007b)
D.C. Douek, J.M. Brenchley, M.R. Betts, D.R. Ambrozak, B.J. Hill, Y. Okamoto, J.P. Casazza, J. Kuruppu, K. Kunstman, S. Wolinsky, Z. Grossman, M. Dybul, A. Oxenius, D.A. Price, M. Connors, R.A. Koup, Nature 417, 95 (2002)
V. Du, O. Yiwen, L. Jingyi, Q. Qiang, A.L. Lindsay, M.H. Doris, P.L. James, Lab Chip 16, 506 (2016)
M.T. Glynn, D. Kinahan, J. Ducree, Lab Chip 14, 2844 (2014)
J.T. Gohring, X. Fan, Sens. Actuators, B 10, 5798 (2010)
X. Jiang, M.G. Spencer, Biosens. Bioelectron. 25, 1622 (2010)
P. Kiesel, M. Beck, N. Johnson, Cytometry, Part A 79, 317 (2011)
G. Kokkinis, S. Cardoso, F. Keplinger, L. Giouroudi, Sens Actuators B Chem. 241, 438 (2017)
C. Kunstmann-Olsen, M.M. Hanczyc, J. Hoyland, S. Rasmussen, H. Rubahn, Sens Actuators B Chem. 229, 7 (2016)
H. Kyungsup, Y.Y. Jin, S. Yong, M.P. Kyong, Lab Chip 16, 132 (2016)
X. Li, C. Breukers, A. Ymeti, K. Pattanapanyasat, K. Sukapirom, L.W. Terstappen, J. Greve, Cytometry, Part B 78, 31 (2010)
H. Masahito, A. Marie, N. Seita, Y. Tomoko, T. Noriyuki, T. Masayuki, N. Satoshi, T. Tsuyoshi, M. Tadashi, Biotechnol. Bioeng. 109, 2017 (2012)
K. Mohammad, D.A. Buchanan, K. Braasch, M. Butler, D.J. Thomson, Sens Actuators B Chem. 249, 246 (2017)
S. Moon, H.O. Keles, A. Ozcan, A. Khademhosseini, E. Hæggstrom, D. Kuritzkes, U. Demirci, Biosens. Bioelectron. 24, 3208 (2009)
A. Ozcan, U. Demirci, Lab Chip 8, 98 (2008)
M.E. Piyasena, S.W. Graves, Lab Chip 14, 1044 (2014)
A. Sposito, V. Hoang, D.L. DeVoe, Lab Chip 16, 3524 (2016)
Z. Wang, S.Y. Chin, C.D. Chin, J. Sarik, M. Harper, J. Justman, S.K. Sia, Anal. Chem. 82, 36 (2009)
N.N. Watkins, S. Sridhar, X. Cheng, G.D. Chen, M. Toner, W. Rodriguez, R. Bashir, Lab Chip 11, 1437 (2011)
C. Willyard, Nat. Med. 13, 1131 (2007)
X. Wu, C.H. Chon, Y.N. Wang, Y. Kang, D. Li, Lab Chip 8, 1943 (2008)
A. Ymeti, X. Li, B. Lunter, C. Breukers, A.G. Tibbe, L.W. Terstappen, J. Greve, Cytometry, Part A. 71, 132 (2007)
H. Yun, H. Bang, J. Min, C. Chung, J.K. Chang, D.C. Han, Lab Chip 10, 3243 (2010)
R. Zachariah, S.D. Reid, P. Chaillet, M. Massaquoi, E.J. Schouten, A.D. Harries, Tropical Med. Int. Health 16, 37 (2011)
H. Zhu, O.I. Serhan, M. Onur, G. Alon, O. Aydogan, Lab Chip 13, 51 (2013)
Acknowledgments
The work was supported by the National Natural Science Foundation of China (No. 81371711) and the research fund to the top scientific and technological innovation team from Beijing University of Chemical Technology (No. buctylkjcx06).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Qiu, X., Yang, S., Wu, D. et al. Rapid enumeration of CD4 + T lymphocytes using an integrated microfluidic system based on Chemiluminescence image detection at point-of-care testing. Biomed Microdevices 20, 15 (2018). https://doi.org/10.1007/s10544-018-0263-y
Published:
DOI: https://doi.org/10.1007/s10544-018-0263-y