Abstract
The rapid, specific and sensitive detection of nucleic acids is of utmost importance for the identification of infectious agents, diagnosis and treatment of genetic diseases, and the detection of pathogens related to human health and safety. Here we report the development of a simple and sensitive nucleic acid sequence-based and Ru(bpy)3 2+-doped silica nanoparticle-labeled lateral flow assay which achieves low limit of detection by using fluorescencent nanoparticles. The detection of the synthetic nucleic acid sequences representative of Trypanosoma mRNA, the causative agent for African sleeping sickness, was utilized to demonstrate this assay. The 30 nm spherical Ru(bpy)3 2+-doped silica nanoparticles were prepared in aqueous medium by a novel method recently reported. The nanoparticles were modified by 3-glycidoxypropyl trimethoxysilane in order to conjugate to amine-capped oligonucleotide reporter probes. The fluorescent intensities of the fluorescent assays were quantified on a mictrotiter plate reader using a custom holder. The experimental results showed that the lateral flow fluorescent assay developed was more sensitive compared with the traditional colloidal gold test strips. The limit of detection for the fluorescent lateral flow assay developed is approximately 0.066 fmols as compared to approximately 15 fmols for the colloidal gold. The limit of detection can further be reduced about one order of magnitude when “dipstick” format was used.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The development of rapid, cost-effective, specific and sensitive methods for the identification and detection of nucleic acids is very important due to their applications in gene expression profiling, and the diagnostics and treatment of clinical diseases. With increasing attention being given to Low-Resource Settings (LRS), new diagnostic methods must be developed which can function with high performance at minimal cost. Agarose, polyacrylamide gel electrophoresis and Western blotting are conventional methods for the detection of nucleic acids, but these methods cannot offer enough specific information and are very laborious and time-consuming (Elbashir et al. 2001; Ganguly et al. 1993). Some methods such as real-time polymerase chain reaction (Jordan and Durso 2005; Vaitilingom et al. 1999), reverse-transcription polymerase chain reaction (Bustin 2000; Pfaffl 2001), and DNA microarrays can provide fast and sensitive identification and detection of the nucleic acid sequences, but these methods need expensive equipment and highly trained personnel. In the past 10 years, some researchers have been focusing on the development of nucleic acid-based biosensors (Barken et al. 2007; Connelly et al. 2008; Lubin and Plaxco 2010). Different labeling and signal amplification techniques were adopted to enhance sensitivities (Shingara et al. 2005; Wang et al. 2001).
Lateral flow assays include both the lateral flow test strip and dipstick formats. While a sample is placed onto the lateral flow assay, the dipstick is inserted into the sample similar to a pregnancy test. Both formats are inexpensive and easy-to-use diagnostic methods which make them ideal for use in LRS such as those affected by Trypanosoma (Yager et al. 2008). Trypanosoma such as Trypanosoma brucei and Trypanosoma cruzi, the causative agents for the potentially fatal African sleeping sickness and Chagas disease, respectively, have important influence on human health. Several approaches are utilized to identify and detect Trypanosoma. Conventional methods such as microscopic examination and xenodiagnoses are considered as standard methods, but they have poor sensitivity and are also labor-intensive and time-consuming (Lane et al. 1997; Reithinger et al. 2010). Immunological methods such as immunochromatographic dipstick test, enzyme-linked immunosorbent assay, immunofluorescence antibody test, and radioimmunosorbent assay are rapid and sensitive but not specific (Kawai et al. 2010; Nieto et al. 2009). Molecular methods such as PCR and real-time nucleic acid sequence-based amplification are very specific but expensive and time consuming, although combination of PCR and chemiluminescence southern blot was used to improve the sensitivity of the detection of Trypanosoma (Chiurillo et al. 2003; Dorn et al. 1999; Gomes et al. 2005; Mohammed et al. 2010; Mugasa et al. 2009; Nantulya 1991; Schapachnik et al. 2009). These techniques are not implemented in Trypanosomiasis control programs due to the high cost of the equipment (Gonzalez et al. 1994; Mugasa et al. 2009). Therefore, there exists a need for an assay which can rapidly, sensitively and specifically detect Trypanosoma without the need for specialized equipment and highly trained personnel.
Colloidal gold and latex particles are commonly used as a marker for lateral flow assays. In addition to gold and latex, other particles such as liposomes have also been investigated to lower the limit of detection (Kumanan et al. 2009; Nugen et al. 2007). We have also previously demonstrated a chemiluminescent lateral flow assay with a low limit of detection (Wang et al. 2012). In this study, a simple, rapid and sensitive fluorescent lateral flow assay by using Ru(bpy)3 2+-doped silica nanoparticle-labeled reporter probe has been developed for nucleic acid sequence-based detection of Trypanosoma mRNA sequences. The reporter and capture probes were designed to target the Trypanosoma mRNA leader sequence, a 35 nucleotide spliced sequence present on the 5’ end of all Trypanosoma mRNA, and the poly A tail found on the 3’ end (Walder et al. 1986). We demonstrate the ability to sensitively detect sub-fmol amount of synthetic leader sequences of Trypanosoma mRNA. The resulting fluorescent lateral flow assay represents an inexpensive, rapid, and sensitive method for Trypanosoma detection without the need for target amplification or costly equipment. For a direct comparison, we have compared the performance of the fluorescent lateral flow with a colloidal gold format.
2 Materials and methods
2.1 Chemicals and materials
Tetraethyl orthosilicate, tris(2,2’-bipyridyl)dichlororuthenium(II) hexahydrate (Ru(bpy)3 2+), and L-lysine were purchased from Sigma-Aldrich (St. Louis, MO, USA). Streptavidin was obtained from SouthernBiotech (Birmingham, AL, USA). Polyvinylpyrrolidone (PVP, molecular weight 8,000) and 3-glycidoxypropyl trimethoxysilane (GOPTMS) were purchased from Acros Organics (NJ, USA). Absolute ethanol, acetone, methanol, toluene, hydrochloric acid, sodium chloride, sodium hydroxide, Triton X-100, and bovine serum albumin were purchased from Fisher Scientific (Fair Lawn, NJ, USA). Sodium phosphate dibasic and borax (sodium tetraborate) were obtained from Sigma-Aldrich Chemical Company (Milwaukee, WI, USA). Sample application pad, nitrocellulose membrane, and absorbent pad were friendly provided by GE Healthcare Bio-Sciences Corp. (Piscataway, NJ, USA). Conjugate release pad was purchased from Life Science-Pall Corp. (NY, USA). The oligonucleotide sequences utilized were purchased from Eurofins MWG Operon (Huntsville, AL, USA) and shown in Table 1. All chemicals used in this study were analytical reagent grade. All solutions and buffers were prepared by using ultrapure water from Barnstead Nanopure InfinityTM ultrapure water system (Thermo Scientific, Dubuque, IA, USA).
2.2 Apparatus
A multi-mode microplate reader (SynergyTM 2) was obtained from BioTek Instruments, Inc. (Winooski, VT, USA). A dispenser (Linomat IV) was purchased from American Laboratory Trading Inc. (East Lyme, CT, USA).
2.3 Preparation of gold nanoparticle-labeled oligonucleotide probe
The mixture of 1625 μL of streptavidin-dressed gold nanoparticles (40 nm, OD520 1.0, BioAssay Works, LLC, MD, USA) and 62.5 μL of 8 μM biotin-capped oligonucleotide (leader sequence complementary, PBS, pH 7.4) were incubated at room temperature for 45 min while shaking. Then 170 μL of 10 % bovine serum albumin with pH 9.0 was added and incubated for another 20 min. The conjugate was centrifuged at 5,000 × g, 4 °C for 20 min. The pellet was resuspended in 2 mM borax, pH 9.0, and adjusted to OD520 3.0.
2.4 Dipstick testing of the target using the gold nanoparticle-labeled oligonucleotide probe
The mixture of 5 μL of the conjugate, 2 μL of the target with the designated concentration, and 3 μL of PBS was incubated in 10 mL glass tube (disposable culture tubes, borosilicate glass, size 10 × 75 mm) at room temperature for 5 min. The test strip was placed into the mixture for 1 min such that the bottom of the strip made contact with the sample. After the incubation step the sample had been completely adsorbed by the test strip. The strip was then moved to another glass tube containing 35 μL PBS and held for 10 min as the PBS pushed the sample past the test and control lines. The strip was taken out from the tube and air-dried for 5 min. The image of the test strip was taken by using a Logitech Webcam and the results were qualitatively determined by eye.
2.5 Preparation of Ru(bpy)3 2+-doped silica nanoparticles
Ru(bpy)3 2+-doped silica nanoparticles were prepared in aqueous medium by using a basic amino acid catalysis method as previously described (Wang et al. 2013). Briefly, tris(2,2’-bipyridyl)dichlororuthenium(II) (Ru(bpy)3 2+) and tetraethyl orthosilicate (TEOS) were added to a reaction system by using three steps in order to control the size of the nanoparticles and the amount of the dye Ru(bpy)3 2+ incorporated inside each nanoparticle. Firstly, 36.1 mg of L-lysine as a basic catalyst was added to 36 ml of deionized water in a polytetrafluoroethylene (PTFE) bottle reactor. After L-lysine was completely dissolved, 0.04 M Ru(bpy)3 2+ solution with the designated volume was added under the agitation to the reactor. Next, TEOS used as a silica source with designated volume was added and mixed in the reactor. Finally, the reactor was put into a 90 °C silicone bath while stirring at 500 rpm. The Ru(bpy)3 2+ solution and the TEOS were added to the system three times in intervals of 24 h. A representative case was the volumes of the Ru(bpy)3 2+ solution/TEOS added are 50 μl/2.7 mL, 400 μl/5.4 mL, and 400 μl/5.4 mL, respectively. After the hydrolysis of TEOS was completed, in order to remove the non-incorporated dye and L-lysine in the solution, orange-colored Ru(bpy)3 2+-doped silica nanoparticle solution was dialyzed in deionized water for 48 h. The deionized water was changed three times during the dialysis. The particles were then concentrated by centrifugation (30,000 × g; 4 °C; 60 min). Finally, the Ru(bpy)3 2+-doped silica nanoparticles were washed twice by absolute ethanol and dried in a vacuum oven (16.8 kPa, 40 °C), which can be used immediately or stored at 4 °C for later usage.
2.6 Surface modification of Ru(bpy)3 2+-doped silica nanoparticles
A portion (2.3 mg) of dried Ru(bpy)3 2+-doped silica nanoparticles (RuSNP) were re-suspended in 4.5 mL dry toluene in a 20 mL glass vial and sonicated at room temperature for 1 h. Then 0.5 mL of GOPTMSwas added to the vial which was placed into a 65 °C water bath while shaking at 200 rpm for 2 h. The pellet was collected following centrifugation (10,000 × g, 4 °C for 30 min), and then sequentially washed in dry toluene, acetone and ethanol, and dried in a vacuum oven overnight (16.8 kPa, 25 °C). The GOPTMS-modified Ru(bpy)3 2+-doped silica nanoparticles (RuSNP-GOPTMS) were immediately used or stored at 4 °C for later usage.
2.7 Preparation of Ru(bpy)3 2+-doped silica nanoparticle-labeled reporter probe
A portion (0.4 mg) of the RuSNP-GOPTMS, 40 μL of 300 μM amine-capped oligonucleotide (reporter), and 280 μL of PBS buffer (40 mmol/L NaCl, 50 mmol/L sodium phosphate buffer, pH 6.8) were incubated at 37 °C for 48 h, while shaking at 800 rpm. Following incubation, the reporter probe- tagged nanoparticles were washed twice with the PBS buffer, and centrifuged at room temperature, 3,500 × g for 4 min. The RuSNP-GOPTMS-oligonucleotide probe was re-suspended in 0.1 mL of the PBS buffer.
2.8 Lateral flow test strip
A lateral flow test strip was prepared according to the literature with modification (Wang et al. 2012). A brief description is as follows:
Immobilization of oligonucleotides on nitrocellulose membrane as capture and control probes
Commercial biotin-capped oligonucleotides such as oligo d(T)25, leader sequence of the target were employed because the oligonucleotides cannot directly be immobilized on the nitrocellulose membrane. Here, the oligonucleotide sequences used as the capture and control probes on the test and control lines were determined on the basis of the sequence of the reporter probe and the target used. One hundred microliter of biotin-oligonucleotide, one hundred microliter of streptavidin, and three hundred microliter of NaHCO3/Na2CO3 buffer (40 mM, pH 9) were mixed and incubated at room temperature. Then the conjugates of the oligonucleotide-biotin-streptavidin were sprayed by a dispenser on the surface of the nitrocellulose membrane (AE 98 Fast, Whatman®) as test line and/or control line, respectively. Oligonucleotides were indirectly immobilized on the nitrocellulose membrane via the interaction among biotin, streptavidin, and the membrane. The distance between a test line and a control line was 8 mm. After vacuum drying at 50 kPa, 40 °C for 1.5 h, the nitrocellulose membrane was blocked using a TBS blocking solution (0.015 % casein sodium salt, 0.3 % polyvinylpyrrolidone, and 0.001 % Tween 20, TBS pH 7.0). After the vacuum drying at 50 kPa, 25 °C for 2 h, the membrane was used immediately or stored at 4 °C.
Preparation of membrane
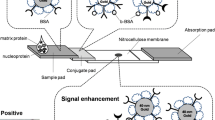
The test strip consists of the sample application pad, conjugate release pad (containing the nanoparticles), nitrocellulose membrane, and absorbent pad, which were all mounted on a plastic backing layer together (Fig. 1). Prior to use, the sample application pad (CF5, Whatman International Ltd.) was saturated with sample pad buffer (0.1 M Na2B4O7.7H2O, pH 8.6; 1 % Triton X-100), and dried at 37 °C overnight and then stored in a desiccator at room temperature until use. The conjugate pad was prepared by immersing the glass fiber pad (Conjugate Pad Grade 8975, Life Science-PALL Corp.) into a conjugate solution (gold-oligonucleotide conjugate with OD520 1.0; 3 % sucrose, PBS, pH 7.4) and drying it at room temperature. Finally, the sample application pad, conjugate release pad, nitrocellulose membrane, and absorbent pad were assembled on a plastic backing using pressure sensitive adhesive tape. The test strips (4 mm wide) were cut using a paper cutter. Each of four components of a lateral flow test strip was overlapped at least 2 mm to make sure the migration of the sample solution through the strip during testing.
2.9 Characterization of various Ru(bpy)3 2+-doped silica nanoparticles
TEM
the un-modified and modified Ru(bpy)3 2+-doped silica nanoparticles in aqueous solutions or their powders were dispersed in the absolute ethanol by sonication for 3 min. The dispersed solutions were dropped onto a copper grid coated with carbon. The particle size, particle size distribution, and morphology of the nanoparticles were measured by transmission electron microscope (TEM, JEOL 100CX, Japan).
Zeta potential
the nanoparticles were re-dispersed in ultrapure water by sonication, and zeta potentials of the nanoparticles were measured by a Zetasizer Nano ZL (Malvern Corporation, United Kingdom).
Excitation wavelength
the nanoparticles were serially diluted in ultrapure water. A fluorescent microtiter plate reader (SynergyTM 2, BioTek Instruments, Inc., Winooski, VT, USA) was used to measure their absorbance spectra from 390 nm to 780 nm with the wavelength interval of 1 nm.
Fluorescence intensity
the nanoparticles prepared by adding different volumes of the Ru(bpy)3 2+ solution were dialyzed by the ultrapure water for 48 h using three water changes. The plate reader was employed to measure the fluorescence intensities of the Ru(bpy)3 2+-doped silica nanoparticles by using excitation wavelength of 460 nm and emission wavelength of 620 nm.
2.10 Fluorescence detection of nucleic acids
For the lateral flow test strip test, 60 μL of target solution with a designated concentration of the Trypanosoma mRNA target was applied to the sample application pad. After the sample has stopped flowing (several minutes), 100 μL deionized water were applied to the sample application pad. The target solution migrated by capillary action through the conjugate release pad, test line, and control line in approximately 10 min. The intensity of the fluorescence signal generated on test and control lines was measured by a fluorescent microtiter plate reader using a custom holder which could fit eight strips per reading (Wang et al. 2012).
For the “dipstick” test, 2.5 μL of the RuSNP-GOPTMS-oligonucleotide conjugate, 2.5 μL of the target with the designated concentration, and 10 μL of PBS or deionized water were incubated in a 10 mL glass tube at room temperature for 5 min while lightly shaking. One end of the test strip (closest to the test line) was soaked into the mixture in the glass tube for 2 min, and then transferred to another glass tube containing 35 μL of PBS or deionized water. Following migration, the strip was taken out from the tube and fixed on the microplate for the fluorescence testing similar the lateral flow test strip.
3 Results and discussion
3.1 Preparation and characterization of Ru(bpy)3 2+-doped silica nanoparticles
The representative TEM image of the spherical Ru(bpy)3 2+-doped silica nanoparticles synthesized by using the novel method is shown in Fig. 2. The size (diameter) of the spherical nanoparticles measured by ImageJ software was 30.9 ± 2.1 nm (Table 2), which is much smaller than that of the Ru(bpy)3 2+-doped silica nanoparticles prepared by using a modified Stöber’s method. The main advantage of this new method compared to the modified Stöber’s method is that the nanoparticles less than 50 nm in size are easily prepared, and the dye content inside the nanoparticles is high, which greatly broadens the applications of the Ru(bpy)3 2+-doped silica nanoparticles, especially in microfluidic and lateral flow biosensors.
The zeta potential of the Ru(bpy)3 2+-doped silica nanoparticles in deionized water was −39.93 ± 1.07 mV, which was very close to that of the bare silica nanoparticles (−39.67 ± 0.98 mV) in deionized water due to the negative charge of hydroxyl groups on the particle surface (Table 2). This demonstrated that the dye Ru(bpy)3 2+ incorporated inside the silica nanoparticles has little influence on the surface of the nanoparticles.
The maximum excited wavelength of the Ru(bpy)3 2+-doped silica nanoparticles was 460 nm. In addition, the nanoparticles have a wide range of the excitation wavelength, e.g. 420 to 500 nm, in which the excitation intensity depends on the concentration of the Ru(bpy)3 2+-doped silica nanoparticles in solution, and the concentration of Ru(bpy)3 2+ within the silica nanoparticles.
3.2 Surface modification of Ru(bpy)3 2+-doped silica nanoparticles by GOPTMS
The schematic diagram of the surface modification of the Ru(bpy)3 2+-doped silica nanoparticles by GOPTMS in toluene is shown in Fig. 3. Dry toluene was used as a solvent in order to form a GOPTMS monolayer on the surface of the Ru(bpy)3 2+-doped silica nanoparticles.
Schematic diagram of the surface modification of Ru(bpy)3 2+-doped silica nanoparticles, the conjugation of oligonucleotide reporter onto the surface of the modified nanoparticles, and the hybridization between the reporter and target. (GOPTMS: 3-glycidoxypropyl trimethoxysilane; RuSNP: Ru(bpy)3 2+-doped silica nanoparticles; LS: leader sequence; LSC: leader sequence complimentary; · Ru(bpy)3 2+ dye molecule)
The size and zeta potential of the nanoparticles in deionized water are shown in Table 2. It can be seen from Table 2 that the size of the GOPTMS-modified nanoparticles is slightly larger than that of the unmodified nanoparticles, which means that following surface modification of the nanoparticles in toluene, a very thin GOPTMS layer was formed. The change in size of the nanoparticles were determined to an extent by the layer numbers of the GOPTMS on the surface of Ru(bpy)3 2+-doped silica nanoparticles. The zeta potential of the GOPTMS-modified nanoparticles in deionized water is very close to neutral, which indicated that the GOPTMS with epoxy group was immobilized onto the surface of the nanoparticles and caused the change of the surface properties of the Ru(bpy)3 2+-doped silica nanoparticles.
3.3 Limit of detection for lateral flow test strip format
The fluorescence intensity on test line as a function of the amount of the nucleic acid target is shown in Fig. 4. The method used for the determination of the limit of detection of the test strip is that the fluorescence intensity of the target on the test line is equal to (m + 3σ), where m and σ are the mean and standard deviation of the fluorescence intensity of the negative control (blank), respectively. The results shown that the limit of detection is 0.066 fmol (corresponding to 1.1 × 10−12 M).
3.4 Limit of detection for “dipstick” format
The dipstick did not contain the sample pad compared with the conventional lateral flow test strip. Here, the main difference in the structure between the “dipstick” format and the lateral flow test strip format is that there is neither sample pad nor conjugate release pad for the “dipstick” format. The limit of detection for “dipstick” format was determined by using the similar approach to the limit of detection for lateral flow test strip format (the fluorescence intensity on test line as a function of the amount of the nucleic acid target was not shown here). The results shown that the limit of detection is approximately 0.004 fmol (corresponding to 1.6 × 10−12 M). Comparing the limits of detection for these two formats it can be seen that the limit of detection of the “dipstick” format on the basis of the amount of the target is six times lower than that of the lateral flow test strip format. As a comparison, the limit of detection for the colloidal gold test strip was approximately 15 fmol (Fig. 5). This demonstrates a significant reduction in the limit of detection when using fluorescence.
The first advantage of the “dipstick” format is that the hybridization between the reporter probe and the target mainly took place off the membrane, which increases hybridization times between the reporter probe and the target. The hybridization with longer time in the tube benefits the decrease of the limit of detection. The second advantage is that no targets are absorbed in the porous sample pad and conjugate release pad, which can reduce the limit of detection to an extent. The absence of sample and conjugate pads means less target will be nonspecifically adsorbed and therefore a higher percentage will make it to the test line. This improved limit of detection is balanced with increased user preparation.
The volumes of the targets used for the testing of “dipstick” and lateral flow test strip formats are 2.5 and 60 μL, respectively, which means 24 times difference. However, the limits of detection on the basis of the concentration of the targets used for these two formats are similar, which means most of the targets in the sample used for testing did not go through the test line or without hybridization between the target and the reporter probe.
The results compare favorable to other novel methods being used for lateral flow such as carbon nanostrings (Kalogianni et al. 2011) and chemiluminescence (Wang et al. 2012) which had limits of detection of 2.5 fmol and 0.5 fmol, respectively.
4 Conclusions
In this study, using Trypanosoma mRNA as a model target, a nucleic acid sequence-based and Ru(bpy)3 2+-doped silica nanoparticles-labeled fluorescent lateral flow assay was developed to detect Trypanosoma. The limits of detection for the lateral flow test strip and “dipstick” formats are approximately 6.6 × 10−2 and 4 × 10−3 fmols, respectively. Based on these results, a general nucleic acid sequence-based and Ru(bpy)3 2+-doped silica nanoparticles-labeled fluorescent lateral flow test strip assay or fluorescent “dipstick” assay was developed to rapidly, sensitively, and specifically test various pathogens and diseases. The most likely reason for the improved limit of detection using the dipstick is that lack of conjugate and sample pad. These pads increase surface area and allow for increased loss of target through nonspecific adsorption. This assay is expected to be used for the rapid, sensitive and specific mass screening of samples and the rapid and accurate detection of the pathogens and diseases so that the prevention, diagnosis and treatments of the diseases are greatly improved.
References
K.B. Barken, J.A.J. Haagensen, T. Tolker-Nielsen, Clin. Chim. Acta 384, 1 (2007)
S.A. Bustin, J. Mol. Endocrinol. 25, 169 (2000)
M.A. Chiurillo, G. Crisante, A. Rojas, A. Peralta, M. Dias, P. Guevara, N. Anez, J.L. Ramirez, Clin. Diagn. Lab. Immunol. 10, 775 (2003)
J.T. Connelly, S.R. Nugen, W. Borejsza-Wysocki, R.A. Durst, R.A. Montagna, A.J. Baeumner, Anal. Bioanal. Chem. 391, 487 (2008)
P.L. Dorn, D. Engelke, A. Rodas, R. Rosales, S. Melgar, B. Brahney, J. Flores, C. Monroy, Am. J. Trop. Med. Hyg. 60, 740 (1999)
S.M. Elbashir, J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, T. Tuschl, Nature 411, 494 (2001)
A. Ganguly, M.J. Rock, D.J. Prockop, Proc. Natl. Acad. Sci. 90, 10325 (1993)
A. Gomes, E. Fernandes, J.L.F.C. Lima, J. Biochem. Biophys. Methods 65, 45 (2005)
N. Gonzalez, I. Galindo, P. Guevara, E. Novak, J.V. Scorza, N. Anez, J.F. Da Silveira, J.L. Ramirez, J. Clin. Microbiol. 32, 153 (1994)
J.A. Jordan, M.B. Durso, J. Mol. Diagn.: JMD 7, 575 (2005)
D.P. Kalogianni, L.M. Boutsika, P.G. Kouremenou, T.K. Christopoulos, P.C. Ioannou, Anal. Bioanal. Chem. 400, 1145 (2011)
M. Kawai, J. Salas, L. Tavares, C. Fontes, J. Infect Dev. Ctries 4, 590 (2010)
V. Kumanan, S.R. Nugen, A.J. Baeumner, Y.F. Chang, J. Vet. Sci. 10, 35 (2009)
J.E. Lane, D. OlivaresVillagomez, C.L. VnencakJones, T.L. McCurley, C.E. Carter, Am. J. Trop. Med. Hyg. 56, 588 (1997)
A.A. Lubin, K.W. Plaxco, Acc. Chem. Res. 43, 496 (2010)
Y.O. Mohammed, M.M. Mohamed-Ahmed, T.K. Lubna, I.E. El Rayah, Afr. J. Biotechnol. 9, 6408 (2010)
C.M. Mugasa, T. Laurent, G.J. Schoone, P.A. Kager, G.W. Lubega, H.D.F.H. Schallig, J. Clin. Microbiol. 47, 630 (2009)
V.M. Nantulya, Experientia 47, 142 (1991)
P.D. Nieto, R. Boughton, P.L. Dorn, F. Steurer, S. Raychaudhuri, J. Esfandiari, E. Gonçalves, J. Diaz, J.B. Malone, Vet. Parasitol. 165, 241 (2009)
S.R. Nugen, B. Leonard, A.J. Baeumner, Biosens. Bioelectron. 22, 2442 (2007)
M.W. Pfaffl, Nucleic Acids Res. 29, e45 (2001)
R. Reithinger, M.J. Grijalva, R.F. Chiriboga, B.A. de Noya, J.R. Torres, N. Pavia-Ruz, P. Manrique-Saide, M.V. Cardinal, R.E. Gürtler, J. Clin. Microbiol. 48, 3003 (2010)
E. Schapachnik, A.R. Riera, S. Dubner, C.F. Filho, A.H. Uchida, C. Ferreira, Cardiol. J. 16, 592 (2009)
J. Shingara, K. Keiger, J. Shelton, W. Laosinchai-Wolf, P. Powers, R. Conrad, D. Brown, E. Labourier, RNA 11, 1461 (2005)
M. Vaitilingom, H. Pijnenburg, F. Gendre, P. Brignon, J. Agric. Food Chem. 47, 5261 (1999)
J.A. Walder, P.S. Eder, D.M. Engman, S.T. Brentano, R.Y. Walder, D.S. Knutzon, D.M. Dorfman, J.E. Donelson, Science 233, 569 (1986)
J. Wang, R. Polsky, D. Xu, Langmuir 17, 5739 (2001)
Y. Wang, C. Fill, S.R. Nugen, Biosensors 2, 32 (2012)
Y. Wang, W. Fan, S.R. Nugen, Mater. Lett. 92, 17 (2013)
P. Yager, G.J. Domingo, J. Gerdes, Annu. Rev. Biomed. Eng. Annu. Rev. Biomed. Eng. 10, 107 (2008)
Acknowledgments
The authors would like to acknowledge USDA special grant #2010-34637-20985 for financial support. This project was supported in part by the UMass Amherst Center for Hierarchical Manufacturing, a nanoscience shared facility funded by the National Science Foundation under NSF Grant #CMMI-1025020.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, Y., Nugen, S.R. Development of fluorescent nanoparticle-labeled lateral flow assay for the detection of nucleic acids. Biomed Microdevices 15, 751–758 (2013). https://doi.org/10.1007/s10544-013-9760-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10544-013-9760-1