Abstract
It is now well-appreciated by philosophers that contemporary large-scale ‘-omics’ studies in biology stand in non-trivial relationships to more orthodox hypothesis-driven approaches. These relationships have been clarified by Ratti (2015); however, there remains much more to be said regarding how an important field of genomics cited in that work—‘genome-wide association studies’ (GWAS)—fits into this framework. In the present article, we propose a revision to Ratti’s framework more suited to studies such as GWAS. In the process of doing so, we introduce to the philosophical literature novel exploratory experiments in (phospho)proteomics, and demonstrate how these experiments interplay with the above considerations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The fields of molecular biology and genetics were transformed upon completion in 2001 of the Human Genome Project (Lander et al. 2001). This provided for the first time near-complete information on the genetic makeup of human beings, and marked the advent of what has become known as the ‘post-genomics’ era, defined by the availability of large-scale data sets derived from ‘genome-scale’ approaches. In turn, this has led to a shift in biological methodology, from carefully constructed hypothesis-driven research, to unbiased data-driven approaches, sometimes called ‘-omics’ studies. These studies have attracted philosophical interest in recent years: see e.g. Burian (2007); O’Malley et al. (2010); Ratti (2015); for more general philosophical discussions of large-scale data-driven approaches in contemporary post-genomics biology, see e.g. Leonelli (2016); Richardson and Stevens (2015).
Recall that -omics studies fall into three main categories: ‘genomics’, ‘transcriptomics’, and ‘proteomics’. The salient features of these three categories as as follows (we make no claim that these features exhaust any of the three categories; they are, however, the features which are relevant to the present article). Genomics is the study of the complete set of genes (composed of DNA) inside a cell. Cellular processes lead to genetic information being transcribed (copied) into molecules known as RNA. ‘Messenger RNA’ (mRNA) carries information corresponding to the genetic sequence of a gene. Transcriptomics is the study of the complete set of RNA transcripts that are produced by the genome. Finally, the information encoded in mRNA is used by cellular machinery called ribosomes to construct proteins; proteomics is the systematic study of these proteins within a cell. Proteins are the ultimate workhorses of the cell; proteomics studies aim to characterise cellular functions mediated by protein networks, in which nodes represent proteins and edges represent physical/functional interactions between them. For further background on genomics, transcriptomics, and proteomics, see Hasin et al. (2017).
Large-scale -omics studies are often described as being ‘hypothesis-free’. To take one example from genomics: advances in genome-editing techniques mean that it is now possible to generate ‘loss-of-function’ mutants in the laboratory. Such mutations are inactivating in the sense that they lead to the loss in the function of a gene within a cell. In the last few years, CRISPR-Cas9 technology has emerged, which makes it possible to create targetted loss-of-function mutants for any of the nearly 20,000 genes in the human genome (Doudna and Charpentier 2014). This allows researchers to ‘screen’ for a gene the loss of which leads to the phenotype of interest, thereby identifying the function of that gene. The methodological idea behind such screening approaches is that one does not require any background hypothesis as to which gene could be involved in a particular biological process, or associated with a particular phenotype: hence the widespread declaration that such approaches are ‘hypothesis-free’ (Shalem et al. 2015). As Burian writes, “Genomics, proteomics, and related “omics” disciplines represent a break with the ideal of hypothesis-driven science” (Burian 2007, p. 289).
With Ratti (2015); Franklin (2005), and others, we find the terminology of ‘hypothesis-free’ to be misleading—for, in fact, such large-scale studies exhibit a Janus-faced dependence on mechanistic hypotheses of a quite standard sort. Ratti characterises such studies, and their connections with more orthodox mechanistic hypothesis-driven science, as involving three steps:
- 1.
The generation of a preliminary set of hypotheses from an established set of premises;
- 2.
The prioritization of some hypotheses and discarding of others by means of other premises and new evidence;
- 3.
The search for more stringent evidence for prioritized hypotheses.
(Ratti 2015, p. 201)
In step (1), scientific hypothesising plays a role, insofar as it is used to delimit the domain of inquiry of the study. For example, a loss-of-function screen to identify the receptor for a pathogen would hypothesise that there exists a non-redundant mechanism for the interaction of the pathogen with the cells, and that the loss of this cellular factor/mechanism would lead to diminution of interaction of the pathogen with the cell surface. For the purpose of the test, such hypotheses are regarded as indubitable: they delimit the range of acceptable empirical enquiry. But there is also a forward-looking dependence of these approaches on scientific hypothesising: the results of such studies can be used to generate more specific mechanistic hypotheses, certain of which are prioritised in step (2) (based on certain additional assumptions—e.g., that there is a single cellular factor/mechanism responsible for pathogen-cell interaction in the above example), and which can then be validated in downstream analysis in step (3). For example, identification of candidate viral receptors using genome-wide loss-of-function screens can be used to generate specific hypotheses regarding the identity of the associated receptor, which can then be subject to empirical test.
Although broadly speaking we concur with Ratti on these matters (in addition to concurring with other philosophers who have written on this topic, e.g. Franklin (2005); Burian (2007)), and find his work to deliver significant advances in our conceptual understanding of such large-scale studies, his citing of ‘genome-wide association studies’ (GWAS) as a means of illustrating the above points (see Ratti 2015, p. 201) invites further consideration. GWAS aims to identify causal associations between genetic variations and diseases/traits; however, it encounters serious difficulties in identifying concrete hypotheses to prioritise, as per Ratti’s (2). Different solutions to this issue (and the related issue of GWAS ‘missing heritability’) manifest in different approaches to this prioritisation: something which deserves to be made explicit in the context of Ratti’s framework. Specifically, while Ratti focuses implicitly on a ‘core gene’ approach to GWAS (cf. Boyle et al. (2017)), according to which a small number of ‘single nucleotide polymorphisms’ (this terminology will be explained in the body of this paper) are primarily responsible for the trait in question (note that this does not imply that only a small number of genes are associated with the relevant phenotype—rather, it assumes that there are some genes which are more central for the manifestation of the phenotype than the majority), there are other approaches to GWAS which do not presuppose this core gene model; as explained in Wray et al. (2018) (albeit without direct reference to Ratti’s work), such approaches would lead to the prioritisation of different hypotheses in Ratti’s (2).Footnote 1
The first goal of the present paper is to expand on these matters in full detail, and to revise Ratti’s framework in order to incorporate the above points: in so doing, we gain a clearer understanding of how GWAS approaches relate to more traditional, mechanistic, hypothesis-driven science. But there is also a second goal of this paper: to explore for the first time (to our knowledge) in the philosophical literature what it would take for the above-mentioned alternative approaches (often relying on network models)—particularly those which appeal to the field of (phospho)proteomics—to succeed. Although we make no claim that such (phospho)proteomics approaches are per se superior to other strategies for hypothesis prioritisation, they are nevertheless in our view worthy of philosophical attention unto themselves, for they constitute (we contend) a novel form of exploratory experimentation (cf. Burian (2007); Franklin (2005); Steinle (1997)) featuring both iterativity (cf. Elliott (2012); O’Malley et al. (2010)) and appeal to deep learning (cf. Bechtel (2019); Ratti (2020)).
Bringing all this together, the plan for the paper is as follows. In Sect. "GWAS studies and prioritisation", we recall the details of GWAS, and witness how different approaches to the so-called missing heritability and coherence problems lead to the prioritisation of different hypotheses in Ratti’s (2). In Sect. "Proteomics and iterative methodology", we turn our attention to network approaches—specifically to those informed by (phospho)proteomics—and study these through the lens of the literature on exploratory experimentation, before returning to our considerations of GWAS and addressing the question of how such network-based approaches inform the question of hypothesis prioritisation in that context. We close with some discussion of future work to be done in the philosophy both of GWAS, and of big-data biology at large.
GWAS studies and prioritisation
Background on GWAS
Many applications of the framework presented in the introduction—perform genome-wide screens based on a general hypothesis (for example, ‘a gene/process is responsible for a disease’), and on the basis of the results obtained construct a more refined hypothesis for further testing—have been highly successful in biomedical research. However, there are cases in which the application of the approach has not been so straightforward. This can best be illustrated using the example of a field of genomics that studies common diseases such as inflammatory bowel disease (IBD), coronary artery disease, insomnia, and depression. These are often diseases complex in nature, and are thought to be controlled not by a single mutation, but rather to be influenced by multiple loci in the genome and even through the effect of the environment.
In the past decades, researchers have developed a method to characterise the genotype-phenotype associations in these diseases: the method is called ‘genome-wide association studies’ (GWAS). To understand this method, it is important to understand single nucleotide polymorphisms (SNPs). SNPs are variations in a single DNA building block, called a ‘nucleotide’, and they constitute the most common type of genetic variation among individuals. There are around 4-5 million SNPs in a person’s genome. Most SNPs have no effect on human health, but there are some cases in which these variations lead to increased chances of disease. GWAS was based originally upon a ‘common disease, common variant’ hypothesis, which states that common diseases can be attributed to common genetic variants (present in more than 1–5% of the population). By scanning the genomes of many different people, GWAS sought to identify the relationships between common genetic variations and common traits. GWAS studies remain very popular in the field of human genetics, and have been successful in identifying a number of novel variant-trait associations (for example, in diseases such as those mentioned above). For a clear introduction to GWAS from the biology literature, see Tam et al. (2019); for existing philosophical works on GWAS, with further details on such studies complimentary to those presented in this paper, see e.g. Bourrat (2020); Bourrat and Lu (2017).
GWAS’ discontents
GWAS is, however, not without its critics. A clear conclusion from multiple GWAS studies is that even statistically highly significant hits identified from such studies are able to account only for a small fraction of the heritability of the trait/disease in question. (Recall that ‘heritability’ is the measure of proportion of the phenotypic variance in a population that can be attributed to genetic differences—see Downes and Matthews (2020) and references therein for further details.) Moreover, GWAS studies often implicate large numbers of genes. To put this into perspective, three GWAS studies performed for height in 2008 identified 27, 12 and 20 associated genomic regions, which accounted merely for 3.7, 2.0, and 2.9% of the population variation in height, respectively ( Lettre et al. (2008); Weedon et al. (2008); Gudbjartsson et al. (2008)). This was in sharp contrast with estimates from previous genetic epidemiology studies, based upon twin studies,Footnote 2 that estimated the heritability of height to be around 80% (Yang et al. (2010)). In the early days of GWAS, this apparent discrepancy from GWAS came to be known as the missing heritability problem. For recent philosophical discussion of this problem, see Bourrat (2020); Bourrat and Lu (2017); Bourrat et al. (2017); Bourrat (2019); Downes and Matthews (2020); Matthews and Turkheimer (2019).
Geneticists have since proposed a number of solutions to the missing heritibility problem. The three most commonly-discussed such solutions are classified by Gibson (2012) as follows:
-
1.
Complex dieseases are polygenic and many loci with small effects account for the phenotype variance.
-
2.
Common diseases are caused by rare genetic variants each of which have large effect sizes.
-
3.
Most common diseases are a result of interactions between many factors such as gene-gene interaction effects and effects from environmental factors.
(We take the proposals for solving the missing heritability problem presented in Bourrat (2020); Bourrat and Lu (2017); Bourrat et al. (2017); Bourrat (2019), which invoke factors from the epigenome, to fall into category (3); we discuss further these proposals in Sect. GWAS reprise.) From multiple GWAS studies on common diseases there is now overwhelming evidence that common diseases are polygenic, as large numbers of genes are often implicated for a given disease. However, using this framework, it is estimated that it would take 90,000–100,000 SNPs to explain 80% of the population variation in height. In light of this, Goldstein (2009) raised the concern with GWAS studies that “[i]n pointing at ‘everything’, the danger is that GWAS could point at ‘nothing”’.
It is understandable that one would find unpalatable its not being the case that a single gene or process can be associated with a particular disease. But the situation here is not as straightforward as the above remarks might suggest. Indeed, Boyle et al. (2017) propose the following refinement of this idea:
Intuitively, one might expect disease-causing variants to cluster into key pathways that drive disease etiology. But for complex traits, association signals tend to be spread across most of the genome—including near many genes without an obvious connection to disease. We propose that gene regulatory networks are sufficiently interconnected such that all genes expressed in disease-relevant cells are liable to affect the functions of core disease-related genes and that most heritability can be explained by effects on genes outside core pathways. We refer to this hypothesis as an ‘omnigenic’ model.
Boyle et al. (2017) propose that within the large number of implicated genes in GWAS, there are a few ‘core’ genes that play a direct role in disease biology; the large number of other genes identified are ‘peripheral’ and have no direct relevance to the specific disease but play a role in general regulatory cellular networks. By introducing their ‘omnigenic’ model, Boyle et al. (2017) acknowledge the empirical evidence that GWAS on complex diseases does in fact implicate large number of genes; they thereby seem to draw a distinction between complex diseases and classical Mendelian disorders, in which small number of highly deleterious variants drive the disease. However, their suggestion of the existence of a small number of ‘core’ genes backtracks on this and paints complex diseases in the same brushstrokes as classical Mendelian disorders. A number of authors have welcomed the suggestion that genes implicated for complex diseases play a role in regulatory networks but have found the dicotomy between core and peripheral genes to be an ill-motivated attempt to fit complex disease into what we intuitively think should be the framework of a disease (‘a small number of genes should be responsible for a given disease’). For example, Wray et al. (2018) write:
It seems to us to be a strong assumption that only a few genes have a core role in a common disease. Given the extent of biological robustness, we cannot exclude an etiology of many core genes, which in turn may become indistinguishable from a model of no core genes.
We concur with this verdict. One possible reconstruction of the reasons underlying the endorsement by Boyle et al. (2017) of ‘core’ versus ’peripheral’ genes could be in order to solve the missing heritability problem. These authors advocate for using experimental methods that are able to identify rare variants that have high effect sizes (solution (2) of the missing heritability problem as presented above), as this is where they suspect the ‘core’ genes can be identified. However, there is at present no evidence that the ‘core gene’ hypothesis need invariably be true for complex diseases (cf. Wray et al. (2018)), so one might be inclined to reject the original hypothesis that all diseases must fit the mould of ‘small number of genes cause complex diseases’. In so doing, one would thereby need to embrace the claim that at least some complex diseases are polygenic and that putative ‘core’ genes are, in fact, no more important than putative ‘peripheral’ genes in this context.
This, however, still leaves us with the original issue that Boyle et al. (2017) were trying to address: how is it that genes which look disconnected are, in fact, together implicated in a given disease? In addressing this question, we again concur with Wray et al. (2018), who write:
To assume that a limited number of core genes are key to our understanding of common disease may underestimate the true biological complexity, which is better represented by systems genetics and network approaches.
That is to say, understanding gene functions and the interplay between the different genes is key to answering why many genes are involved in complex diseases. This is not a straightforward task and a full characterisation of the roles that genes play in biological systems remains a distant prospect.
One approach to addressing this issue is to identify relationships between genes in a cell by way of a systems biology approach, underlying premises of which are that cells are complex systems and that genetic units in cells rarely operate in isolation. Hence, on this view, understanding how genes relate to one another in a given context is key to establishing the true role of variants identified from GWAS hits. There are a number of approaches described in the field of systems biology to identify gene-gene relationships. One widely-implemented approach is to construct ‘regulatory networks’ relating these genes. A regulatory network is a set of genes, or parts of genes, that interact with each other to control a specific cell function. With recent advances in high-throughput transcriptomics, it is now possible to generate complex regulatory networks of how genes interact with each other in biological processes and define the roles of genes in a context-dependent manner based on mRNA expression in a cell. As the majority of GWAS hits often lie in non-coding regions of the genome, which are often involved in regulating gene expressions, networks based on mRNA expression are powerful means to interpret of the functional role of variants identified by GWAS.
Another approach to the functional validation of GWAS hits—currently substantially less common—proceeds by constructing networks generated from expression of proteins/phosphoproteins in a cell (more details of these approaches will be provided in the following section). Such approaches would in principle depict completely the underlying state of the cell. Combined with gene expression data, protein expression networks and signalling networks from proteomics would make transparent the functional role of the variants identified in GWAS studies in a given context—that is, they would provide a mechanistic account of disease pathogenesis without recourse to a neo-Mendelian ‘core gene’ model. Genes which prima facie appear disconnected and irrelevant to disease biology may be revealed by these approaches to be relevant after all. To illustrate, consider a complex disease such as IBD: it is thought that both (i) a disturbed interaction between the gut and the intestinal microbiota, and (ii) an over-reaction of the immune system, are required for this disease phenotype to manifest. Thus, it is likely that a number of genetic pathways will be important—pathways which need not prima facie be connected, but which may ultimately be discovered to be related in some deeper way. These proteomics-informed network approaches would thereby afford one resolution to what has been dubbed by Reimers et al. (2019) and Craver et al. (2020) the ‘coherence problem’ of GWAS: to explain how it is that all genes implicated in these studies are related to one another mechanistically.Footnote 3 Clearly, these approaches could be brought to bear in order to vindicate responses (1) or (3) to the missing heritability problem, presented above.Footnote 4
To close this subsection, it is worth reflecting on how the ‘core gene’ hypothesis might intersect with network-based approaches. If a core gene exists, then a network analysis should (at least in principle) be able to identify it; in this sense, a ‘core gene’ hypothesis can be compatible with a network approach. As already mentioned above, however, there is no evidence that such core genes invariably exist: a network analysis could (in principle) identify many ‘central hubs’, rather than just one—an outcome not obviously compatible with the ‘core gene’ hypothesis. (For more on this latter possibility, cf. the very recent work of Barrio-Hernandez et al. (2021), discussed further below.)
Ratti’s framework for large-scale studies
Suppose that one follows (our reconstruction of) Boyle et al. (2017), in embracing option (2) presented above as a solution to the GWAS missing heritability problem. One will thereby, in Ratti’s second step in his three-step programme characterising these data-driven approaches to biology, prioritise hypotheses according to which a few rare genes are responsible for the disease in question. This, indeed, is what Ratti (2015) suggests in §2.2 of his article. However, one might question whether this prioritisation is warranted, in light of the lack of direct empirical evidence for this neo-Mendelian hypothesis (as already discussed). Wray et al. (2018), for example, write that
... [t]o bias experimental design towards a hypothesis based upon a critical assumption that only a few genes play key roles in complex disease would be putting all eggs in one basket.
If one concurs with Wray et al. (2018) on this matter (as, indeed, we do), then one may prioritise different hypotheses in the second step of Ratti’s programme—in particular, one may prioritise specific hypotheses associated with ‘polygenic’ models which would constitute approach (1) and/or approach (3) to the missing heritability problem.
This latter point should be expanded. Even if one does embrace a ‘polygenic’ approach to the missing heritability problem (i.e., approach (1) and/or approach (3)), and applies e.g. networks (whether transcriptomics-based, or (phospho)proteomics-informed, or otherwise—nothing hinges on this for our purposes here) in order to model the genetic factors responsible for disease pathogenesis, ultimately one must prioritise specific hypotheses for laboratory test. For example, Schwartzentruber et al. (2021) implement in parallel a range of network models within the framework of a polygenic approach in order to prioritise genes such as TSPAN14 and ADAM10 in studies on Alzheimer’s disease (we discuss further the methodology of Schwartzentruber et al. (2021) in §3.3). Note, however, that these specific hypotheses might be selected for a range of reasons—e.g., our prior knowledge of the entities involved, or ease of testability, or even financial considerations—and that making such prioritisations emphatically does not imply that one is making implicit appeal to a ‘core gene’ model. This point is corroborated further by the fact that the above two genes are not the most statistically significant hits in the studies undertaken by Schwartzentruber et al. (2021), as one might expect from those working within the ‘core gene’ framework.
Returning to Ratti’s framework: we take our noting this plurality of options vis-à-vis hypothesis prioritisation to constitute a friendly modification to this framework appropriate to contexts such as that of GWAS. But of course, if one were to leave things here, questions would remain—for it would remain unclear which polygenic model of disease pathogenesis is to be preferred, and how such models are generated. Given this, it is now incumbent upon us to consider in more detail how such approaches set about achieving these tasks in practice: due both to their potential to offer underlying mechanistic models of the cell, as well as due to the novel iterative methodology for hypothesis generation involved, we focus largely in the remainder upon (phospho)proteomics-based approaches.
Proteomics and iterative methodology
Proteomics promises to afford the ultimate fundamental mechanistic account of cellular processes; data from proteomics would, therefore, illuminate the underlying relationships between the variants identified in GWAS studies. In this section, we explore in greater detail how such proteomics approaches proceed; they constitute a novel form of ‘exploratory experimentation’ (in the terminology of Burian (2007); Steinle (1997)) worthy unto themselves of exposure in the philosophical literature.Footnote 5 In proteomics, further complications for hypothesis generation and testing arise, for data is sparse, and experiments often prohibitively expensive to perform. Given these constraints, how is progress to be made? It is to this question which we now turn; the structure of the section is as follows. In Sect. Proteomics: a data-deprived field, we present relevant background regarding proteomics. Then, in Sect. Methodological iteration, we argue that the development of this field can be understood on a model of a novel form of iterative methodology (cf. Chang 2004; O’Malley et al. 2010). We return to the relevance of these approaches for GWAS in Sect. GWAS reprise.
Proteomics: a data-deprived field
The ultimate aim of -omics studies is to understand the cell qua biological system. Transcriptomics is now sufficiently well-advanced to accommodate large-scale systematic studies to the point of being used to validate variants identified from GWAS.Footnote 6 By contrast, proteomics—the study of proteins in a cell—remains significantly under-studied. Technologies allowing for the systematic study of proteins are not as advanced as those for studying genes and transcripts; this is mainly because no method currently exists for directly amplifying proteins (i.e., increasing the amount of a desired protein in a controlled laboratory context): a methodology which has been key for genomics and transcriptomics. Proteins are very diverse in the cell: a single gene/transcript gives rise to multiple proteins. Proteins themselves can be modified in the cell after being created, thus further increasing the complexity of proteomics studies. Unlike genomics and transcriptomics, in which it is now common to perform systematic genome-wide or transcriptome-wide approaches, studies of proteins are therefore usually taken piecemeal.
Proteomics research tends to focus on families of proteins that are involved in a particular known biological process. Among the important families of proteins are kinases and phosphatases, which are molecules that are responsible for signal transmission in the cell. These proteins are able to modify other proteins by adding or removing a phosphate group (respectively). This modification changes the shape (‘conformation’) of the protein, rendering it active or inactive,Footnote 7 depending on the context. By examining the phopsphorylation state of the proteins inside a cell, it is possible to infer the signalling state of that cell. The field of phosphoproteomics aims to characterise all phospho-modified proteins within a cell. This is thought to be one of the most powerful and fundamental ways of inferring the signalling process within a cell; the approach could add a substantial new layer to our understanding of both basic and disease biology. That said, a recent estimate suggests that current approaches have identified kinases for less than 5% of the phosphoproteome. What is even more staggering is that almost 90% of the phosphorylation modifications that have been identified have been attributed to only 20% of kinases. The other 80% of the kinases are completely dark: their functions remain unknown. For many such kinases, we do not even know where in the cell they are located. (See Needham et al. (2019) for a review of the current state of play in phosphoproteomics.)
In such a field, systematic studies to quantify the entire phosphoproteome in a cell and an ability to assign a kinase to every phosphorylated component would be the ultimate aim. But phosphoproteomics studies themselves are currently extremely expensive, and there are technological limitations in mapping the global phosphoproteome—not least sparsity of data, which often comes as a result of limitations in the technical setup of laboratory measurements and experiments. For example: the same sample measured in the same machine at two different instances will give readings for different phosphoproteins. Some statistical methods can be used to overcome these limitations, but these require making assumptions regarding the underlying biology, which defeats the point of an unbiased study.
In spite of these difficulties, it has been shown that if one combines multiple large-scale phosphoprotemics data sets (each admittedly incomplete), it is possible to predict kinase-kinase regulatory relationships in a cell using data-driven phosphoprotein signalling networks obtained via supervised machine learning approaches (a recent study from Invergo et al. 2020 showcases one such approach; we will use this as a running example in the ensuing).Footnote 8 First, a training set of data is used to teach a machine a classification algorithm. Once the classification algorithm is learnt, the machine is set to the task of applying it to unlabelled data: in our case, the goal is to identify further, as-yet unknown, regulatory protein relationships or non-relationships. (On machine learning and network analysis of biological systems, see also Bechtel (2019) and Ratti (2020).)
Before assessing such phosphoproteomics machine learning algorithms as that of Invergo et al. (2020), there are two further complications with the current state of play in proteomics which need to be mentioned. First: it is much easier to curate positive lists of interactions than negative lists. (This is essentially a case of its being easier to confirm existentially quantified statements than universally quantifies statements: for how can we ever truly ascertain that any two given proteins never interact?) Thus, at present, negative lists obtained from laboratory experiments are underpopulated. Invergo et al. (2020) attempt to circumvent this issue in the following way: they assume that regulatory relationships are rare, so that if one were to randomly sample protein associations, one could create reliably large artificial negative sets; indeed, they do generate artificial negative sets in exactly this way. (Clearly, this means that these approaches again cannot be understood as being ‘hypothesis-free’: cf. Sect. Introduction.)
The second problem with the current state of play in proteomics is this: when a given interaction occurs is a function of multifarious factors, most notably cell context. This context-dependence means that an entry in a negative set in one context might, in fact, be an entry in a positive set in another. To illustrate: in the case of regulatory relationships between two kinases, it is known that such relationships can be prone to dysregulation in diseases such as cancer. Hence, a well-annotated positive set relationship can very well be dysregulated in a cancer context, so that this relationship no longer exists, effectively putting it into a negative set. The problem is that many data-driven approaches rely on data that are generated in simple reductionist systems such as cancer cell lines—so that the results obtained might not carry across to the target physiological context. (Cancer cell lines can grow infinitely, and thus are ideal for experiments.) The approach taken by Invergo et al. (2020) utilises data from breast cancer cell lines; hence, the relationships they predict could be specific to a dysregulated system. In response to this second problem, we suggest replying on behalf of Invergo et al. (2020) that most regulatory relationships fundamental to the functioning of the cell should hold true in most contexts. At present, however, given the data-deprived nature of proteomics, there is little direct evidence for this hypothesis. (Again, the appeal to any such hypothesis would mean that such proteomics approaches cannot be ‘hypothesis-free’.)
Thus, the fact that Invergo et al. (2020) utilise data from breast cancer cell lines raises the possibility that their machine learning algorithms might be trained on data unsuited to other contexts, leading to concerns regarding error propagation. This general concern regarding the context-specificity (or lack thereof) of input data sets is, however, recognised by authors in the field—for example, Barrio-Hernandez et al. (2021) note that “improvements in mapping coverage and computational or experimental approaches to derive tissue or cell type specific networks could have a large impact on future effectiveness of network expansion” (Barrio-Hernandez et al. 2021, p. 14).
Methodological iteration
In spite of these problems, Invergo et al. (2020) argue that the results obtained from their approach afford a useful means of bootstrapping further progress in phosphoproteomics. As they put it:
Although we do not suggest that these predictions can replace established methods for confirming regulatory relationships, they can nevertheless be used to reduce the vast space of possible relationships under consideration in order to form credible hypotheses and to prioritize experiments, particularly for understudied kinases. (Invergo et al. 2020, p. 393)
One way to take this point is the following. Ideally, in order to construct positive and negative sets, one would test in the laboratory each individual protein association. Practically, however, this would be an unrealistic undertaking, as we have already seen. What can be done instead is this:
-
1.
Generate a global phosphoproteomics data set, albeit one that is incomplete and sparse (e.g., that presented in Wilkes et al. (2015)), based upon laboratory experiments.
-
2.
Train, using this data set and input background hypotheses of the kind discussed above, a machine learning algorithm (such as that presented in Invergo et al. (2020)) to identify candidate interactions in the unknown space of protein-protein interactions.Footnote 9
-
3.
Use these results to guide further laboratory experimentation, leading to the development of more complete data sets.
-
4.
Train one’s machine learning algorithms on these new data sets, to improve performance; in turn, repeat further the above process.
Clearly, a process of reflective equilibrium is at play here (cf. Daniels (2016)). As is well-known, Chang (2004) has proposed an iterative conception of scientific methodology, according to which the accrual of scientific hypotheses is not a linear matter; rather, initial data may lead to the construction of a theoretical edifice which leads one to develop new experiments to revise one’s data; at which point, the process iterates. This fits well with the above-described procedures deployed in phosphoproteomics; it also accords with previous registration of the role of iterative procedures in large-scale biological studies—see e.g. O’Malley et al. (2010) and Elliott (2012).
Let us delve into this a little deeper. As Chang notes,
There are two modes of progress enabled by iteration: enrichment, in which the initially affirmed system is not negated but refined, resulting in the enhancement of some of its epistemic virtues; and self-correction, in which the initially affirmed system is actually altered in its content as a result of inquiry based on itself. (Chang 2004, p. 228)
Certainly and uncontroversially, enrichment occurs in the above four-step process in phosophoproteomics: the new data yield a refinement of our previous hypotheses in the field. In addition, however, it is plausible to understand the above iterative methodology as involving self-correction: for example, in might be that the machine learning algorithm of Invergo et al. (2020) identifies a false positive, yet nevertheless makes sufficiently focused novel predictions with respect to other candidate interactions in order to drive new experimentation, leading to a new data set on which the algorithm can be trained, such that, ultimately, the refined algorithm does not make a false positive prediction for that particular interaction. This is entirely possible in the above iterative programme; thus, we maintain that both modes of Changian iterative methodology are at play in this approach.
There is another distinction which is also relevant here: that drawn by Elliott (2012) between ‘epistemic iteration’—“a process by which scientific knowledge claims are progressively altered and refined via self-correction or enrichment”—and ‘methodological iteration’—“a process by which scientists move repetitively back and forth between different modes of research practice” (Elliott 2012, p. 378). It should be transparent from our above discussion that epistemic iteration is involved in these proteomics approaches. Equally, though, it should be clear that methodological iteration is involved, for the approach alternates between machine learning and more traditional laboratory experimentation. That machine learning can play a role in an iterative methodology does not seem to have been noted previously in the philosophical literature—for example, it is not identified by Elliott (2012) as a potential element of a methodologically iterative approach; on the other hand, although the role of machine learning in network modelling and large-scale studies is acknowledged by Bechtel (2019) and Ratti (2020) (the latter of whom also discusses—albeit without explicitly using this terminology—the role of machine learning in epistemic iteration: see (Ratti 2020, p. 89)), there is no mention of its role in an iterative methodology such as that described above.
GWAS reprise
Given the foregoing, we hope it is reasonable to state that the approaches to proteomics of e.g. Invergo et al. (2020) constitute novels forms of exploratory experimentation, worthy of study in their own right. Let us, however, return now to the matter of polygenic approaches to GWAS hits. In principle, the results of the methodologies of e.g. Invergo et al. (2020) could further vindicate these approaches, by providing mechanistic models of which genes interact in a disease context, and when and why they do so. In turn, they have the capacity to allow biologists to prioritise specific hypotheses in Ratti’s step (2), without falling back upon assumptions that only few genes are directly involved in complex disease biology.
Note that that there is a complex interplay between this iterative methodology and the ‘eliminative induction’ of stages (1) and (2) Ratti’s analysis (see Sect. Introduction; for earlier sources on eliminative induction, see Earman (1992); Kitcher (1993); Norton (1995)). We take this to consist in the following. First, a methodology such as that of Invergo et al. (2020) is used to generate a particular network-based model for the factors which are taken to underlie a particular phenotype. This model is used to prioritise (à la eliminative induction) particular hypotheses, as per stage (2) of Ratti’s framework; these are then subject to specific test, as per stage (3) of Ratti’s framework. The data obtained from such more traditional experimentation is then used to construct more sophisticated network models within the framework of Invergo et al. (2020); these in turn lead to the (eliminative inductive) prioritisation of further specific hypotheses amenable to specific test. As already discussed above, this is a clear example of the ‘methodological iteration’ of Elliott (2012).
It bears stressing that (phospho)proteomics network-based approaches may, ultimately, constitute only one piece of the solution to the broader puzzle that is GWAS hypothesis prioritisation. In very recent work, Schwartzentruber et al. (2021) have brought to bear upon this problem consideration of, inter alia, epigenomic factors alongside network-based analyses. There are two salient points to be made on this work. First: although Bourrat et al. (2017) are correct that epigenomic studies and background may have a role to play in addressing the missing heritability problem (cf. Bourrat (2019, 2020); Bourrat and Lu (2017)), a view in contemporary large-scale biological studies—evident in papers such as Schwartzentruber et al. (2021)—is that these considerations can be supplemented with yet other resources, such as network-based studies; we concur with this verdict. Second: in order to construct these networks, Schwartzentruber et al. (2021) rely on established protein-protein interaction databases such as STRING, IntAct and BioGRID (Schwartzentruber et al. 2021, p. 397). While effective in their own right, networks developed from such databases have the disadvantage that they represent signalling in an ‘average’ cell, and are therefore unsuitable for studying dynamic context- and cell-type-specific signalling responses (cf. Sharma and Petsalaki (2019)). In this regard, it would (at least in principle) be preferable to utilise regulatory and context-specific networks developed using methods described in work such as that of Invergo et al. (2020) in future approaches to GWAS hypothesis prioritisation. That being said, in practice this may not yet be fruitful, as at present contemporary large-scale biology is only at the early stages of the iterative processes discussed above; moreover, the training data sets used by such methods remain at this stage not completely context-specific (recall that Invergo et al. (2020) utilise a breast cancer training set)—meaning that the potential of such work to yield detailed, context-specific network-based models is yet to be realised in full.
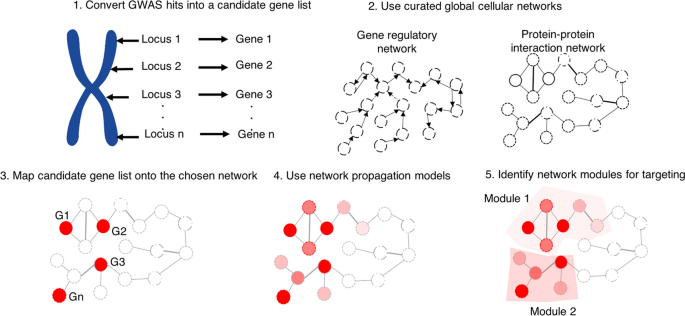
With all of the above in hand, we close this subsection by considering more precisely the question of how the machine learning algorithms of Invergo et al. (2020) bear upon the missing heritability problem. Having developed regulatory protein-protein interaction networks on the basis of such algorithms, one can take (following here for the sake of concreteness the lead of Barrio-Hernandez et al. (2021)) the connection with hypothesis prioritisation in GWAS (and, in turn, the missing heritability problem) to proceed via the following steps (also summarised visually in Fig. 1):
-
1.
Select a protein-protein interaction network. Usually, this is a pre-existing curated network, such as those defined in the STRING database (discussed above). However, instead of such curated networks, use in their place networks developed on the machine learning models of e.g. Invergo et al. (2020).
-
2.
Within those networks, identify the nodes (i.e., proteins) which correspond to hits from a particular GWAS (i.e., the proteins associated with the genes identified in the GWAS).Footnote 10
-
3.
Use network propagation methods (see e.g. Cowen et al. (2017) for a review of such methods), potentially alongside other factors (as discussed in e.g. Schwartzentruber et al. (2021)) in order to identify known modules (i.e., separated substructures within a network) associated with the disease in question.
-
4.
Target elements of those modules, regardless of whether or not they were hits in the original GWAS. (This latter approach—of targeting beyond the original GWAS hits—is novel to the very recent work of Barrio-Hernandez et al. (2021).)
The application of networks to GWAS hit prioritisation. In (1), GWAS hits are converted to candidate gene lists. In (2), one selects a cellular network: this could be a gene regulatory network, or a protein-protein interaction network (e.g. from STRING), or a protein-protein regulatory network (possibly constructed via the machine learning methodologies of Invergo et al. (2020)). In (3), genes associated with the GWAS loci are mapped to the chosen network. In (4), network propagation methods (e.g. diffusion techniques) are applied in order identify potential disease-related genes not picked up by the GWAS. In (5), the results of these network analyses are used to identify significant genetic modules to be targeted experimentally in investigations into disease pathogenesis. Note, following Wray et al. (2018) and Barrio-Hernandez et al. (2021), that this particular means of bridging the gap between cellular networks and investigations into the results of GWAS hits does not presuppose a ‘core gene’ hypothesis
On (2) and (3): Boyle et al. (2017) may or may not be correct that many genes are implicated (either in the original screen, or after the network analysis has been undertaken)—recall from Sect. GWAS’ discontents their ‘omnigenic’ model. However, on the basis of the work of Barrio-Hernandez et al. (2021) one might argue that this is not the most important question—rather, the important question is this: which gene modules provide insights into the disease mechanism? One can ask this question without subscribing to a ‘core gene’ model; thus, we take the work of Barrio-Hernandez et al. (2021) to be consistent with the above-discussed points raised by Wray et al. (2018).
Outlook
This paper has had two goals. The first has been to propose revisions to the framework of Ratti (2015) for the study of the role of hypothesis-driven research in large-scale contemporary biological studies, in light of studies such as GWAS and its associated missing heritability problem. In this regard, we have seen that different hypotheses may be prioritised, depending upon whether one adopts a ‘core’ gene model (as Ratti (2015) assumes, and as is also advocated in Boyle et al. (2017)), or whether one adopts a polygenic model (as endorsed by Wray et al. (2018); cf. Barrio-Hernandez et al. (2021)). The second goal of this paper has been to consider how these hypotheses would be developed on polygenic approaches via (phospho)proteomics—which itself constitutes a novel form of exploratory experiment, featuring as it does both iterativity and deep learning—and to consider what it would take for these network-based proteomics approaches to succeed. A broader upshot of this paper has been the exposure for the first time to the philosophical literature of proteomics: given its potential to provide mechanistic models associated with disease phenotypes, the significance of this field cannot be overstated.
The issues discussed in this paper raise important questions regarding how researchers prioritise not just first-order hypotheses as per Ratti’s (2), but also the background assumptions which allow one to make such adjudications to begin with. To be concrete: in the case of GWAS, should one prioritise the assumption that rare variants of large effect in a small number of genes drive complex diseases, or rather invest in developing systems-based approaches and in improving under-studied fields, such as (phospho)proteomics, which may or may not ultimately shed light on the question of why complex diseases have thus far manifested empirically as polygenic? These choices lead to different first-order prioritisations in Ratti’s second step, and thereby have great potential to steer the course of large-scale studies in future years. Given limited resources in the field, it is, in our view, worth pausing to reflect on whether said resources are appropriately allocated between these options, and to strive to avoid any status quo bias in favour of currently-popular assumptions.Footnote 11
Notes
Twin studies are powerful approaches to studying the genetics of complex traits. In simple terms, twin studies compare the phenotypic similarity of identical (monozygotic) twins to non-identical (dizygotic) twins. As monozygotic twins are genetically identical and non-identical twins are on average ‘half identical’, observing greater similarity of identical over non-identical twins can be used as an evidence to estimate the contribution of genetic variation to trait manifestation. For further discussion of twin studies in the philosophical literature, see e.g. Matthews and Turkheimer (2019); Downes and Matthews (2020).
There are many further questions to be addressed here in connection with the literature of mechanisms and mechanistic explanations. For example, are these network approaches best understood as revealing specific mechanisms, or rather as revealing mechanism schema (to use the terminology of (Craver and Darden 2013, ch.3))? Although interesting and worthy of pursuit, for simplicity we set such questions aside in this paper, and simply speak of certain contemporary biology approaches as revealing ‘underlying mechanisms’. In this regard, we follow the lead of Ratti (2015).
To be completely clear: we do not claim that these (phospho)proteomics-based network approaches are superior to regulatory network approaches, given the current state of technology in the field. On the contrary—as we explain in Sect. Proteomics and iterative methodology—the former of these fields is very much nascent, and has yet to yield significant predictive or explanatory fruit. Nevertheless—again as we explain in Srct. Proteomics and iterative methodology—in our view these approaches are worthy of exposure in the philosophical literature in their own right, for (a) they offer one of the most promising means (in principle, if not yet in practice) of providing a mechanistic account of disease pathogenesis, and (b) the particular way in which hypotheses are developed and prioritised on these approaches is conceptually rich.
Recall: “Experiments count as exploratory when the concepts or categories in terms of which results should be understood are not obvious, the experimental methods and instruments for answering the questions are uncertain, or it is necessary first to establish relevant factual correlations in order to characterize the phenomena of a domain and the regularities that require (perhaps causal) explanation” (Burian 2013). Cf. e.g. Franklin (2005); Steinle (1997). All of the -omics approaches discussed in this paper were identified in Burian (2007) as cases of exploratory experimentation; the details of contemporary proteomics approaches have, however, not been presented in the philosophical literature up to this point (at least to our knowledge).
In this paper, we do not go into the details of specific transcriptomics studies. One interesting approach worthy of mention, however, is ‘single-cell RNA sequencing’ (SC-RNA), which allows biologists to assay the full transcriptome of hundreds of cells in an unbiased manner (see e.g. Hwang et al. (2018) for a recent review). The advantage of SC-RNA over older methods lies in its ability to identify the transcriptomes from heterocellular and poorly-classified tissue populations and disease-associated cell states.
As the addition or removal of phosphate groups regulates the activity of a protein, such relationships between a kinase and its target (also called a ‘substrate’) are referred to as ‘regulatory relationships’. Kinases themselves can also be phosphorylated by other kinases, so there exist also kinase-kinase regulatory relationships in a cell.
Supervised machine learning involves training a machine on a given data set (for example, a collection of cat photos versus dog photos), before assigning the machine the task of classifying entries in some new data set. By contrast, in unsupervised learning, the machine is instructed to find its own patterns in a given data set. For some recent philosophical considerations regarding machine learning, see Sullivan (2019).
One can also test the results of the machine binary classification algorithm on other data sets: this Invergo et al. (2020) did with reference to the data presented in Hijazi et al. (2020). The design of the algorithmic system and algorithm used by Invergo et al. (2020) is described with admirable clarity at (Invergo et al. 2020, pp. e5ff.), to which the reader is referred for further details.
Note that identification of candidate genes from the loci which constitute GWAS hits is non-trivial. The recently-described ‘locus-to-gene’ (L2G) approach is a machine learning tool which can be used to prioritise likely causal genes at each locus given genetic and functional genomics features (see Mountjoy et al. (2020)).
References
Barrio-Hernandez I, Schwartzentruber J, Shrivastava A, del Toro N, Zhang Q, Bradley G, Hermjakob H, Orchard S, Dunham I, Anderson CA, Porras P, Beltrao P (2021) ‘Network expansion of genetic associations defines a pleiotropy map of human cell biology’, bioRxiv. https://www.biorxiv.org/content/early/2021/07/19/2021.07.19.452924
Bechtel W (2019) Hierarchy and levels: analysing networks to study mechanisms in molecular biology. Philos Transact R Soc B 375(20190320):20190320
Bourrat P (2019) Evolutionary transitions in heritability and individuality. Theory Biosci 138:305–323
Bourrat P (2020) Causation and single nucleotide polymorphism heritability. Philos Sci 87:1073–1083
Bourrat P, Lu Q (2017) Dissolving the Missing Heritability Problem. Philosophy of Science 84:1055–1067
Bourrat P, Lu Q, Jablonka E (2017) Why the missing heritability might not be in the DNA. BioEssays 39:1700067
Boyle E, Li Y, Pritchard J (2017) An expanded view of complex traits: from polygenic to omnigenic. Cell 169:1177–1186
Burian R (2013) Exploratory experimentation. In: Dubitzky W, Wolkenhauer O, Cho K-H, Yokota H (eds) Encyclopedia of systems biology. Springer, Berlin
Burian RM (2007) On MicroRNA and the need for exploratory experimentation in post-genomic molecular biology. Hist Philos Life Sci. 29(3):285–311. http://www.jstor.org/stable/23334263
Chang H (2004) Inventing temperature: measurement and scientific progress. Oxford University Press, Oxford
Cowen L, Ideker T, Raphael BJ, Sharan R (2017) Network propagation: a universal amplifier of genetic associations. Nat Rev Genet 18(9):551–562. https://doi.org/10.1038/nrg.2017.38
Craver CF, Darden L (2013) In search of mechanisms. University of Chicago Press, Chicago
Craver CF, Dozmorov M, Reimers M, Kendler KS (2020) Gloomy prospects and roller coasters: finding coherence in genome-wide association studies. Philos Sci 87(5):1084–1095
Daniels N (2016) Reflective equilibrium. The Stanford Encyclopedia of Philosophy
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096
Downes SM, Matthews L (2020) Heritability. In: Zalta EN (ed) The Stanford encyclopedia of philosophy. Stanford University, Metaphysics Research Lab
Earman J (1992) Bayes or bust? a critical examination of BayesianConfirmation Theory,. MIT Press, Cambridge
Elliott KC (2012) Epistemic and methodological iteration in scientific research. Stud Hist Philos Sci Part A 43(2):376–382
Franklin L (2005) Exploratory experiments. Philos Sci. 72(5):888–899. https://www.jstor.org/stable/10.1086/508117
Gibson G (2012) Rare and common variants: twenty arguments. Nat Rev Genet 13(2):135–145
Goldstein D (2009) Common genetic variation and human traits. N Engl J Med 360:1696–1698
Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, Helgadottir A, Ingason A, Steinthorsdottir V, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Pedersen O, Aben KK, Witjes JA, Swinkels DW, Heijer Md, Franke B, Verbeek ALM, Becker DM, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Gulcher J, Kiemeney LA, Kong A, Thorsteinsdottir U, Stefansson K (2008) Many sequence variants affecting diversity of adult human height. Nat Genet 40(5):609–615. https://doi.org/10.1038/ng.122
Hasin Y, Seldin M, Lusis A (2017) Multi-omics approaches to disease. Genome Biol 18(1):83
Hijazi M, Smith R, Rajeeve V, Bessant C, Cutillas PR (2020) Reconstructing kinase network topologies from phosphoproteomics data reveals cancer-associated rewiring. Nat Biotechnol 38(4):493–502
Hwang B, Lee JH, Bang D (2018) Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 50(8):96. https://doi.org/10.1038/s12276-018-0071-8
Invergo BM, Petursson B, Akhtar N, Bradley D, Giudice G, Hijazi M, Cutillas P, Petsalaki E, Beltrao P (2020) Prediction of signed protein kinase regulatory circuits. Cell Syst 10(5):384-396.e9
Kitcher PS (1993) The advancement of science. Oxford University Press, Oxford
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts, P, Koonin, E V, Korf I, Kulp, D, Lancet D, Lowe T M, McLysaght A, Mikkelsen T, Moran J V, Mulder N, Pollara V J, Ponting C P, Schuler G, Schultz J, Slater G, Smit A F, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf Y I, Wolfe, K H, Yang S P, Yeh R F, Collins F, Guyer M S, Peterson J, Felsenfeld A, Wetterstrand K A, Patrinos A, Morgan M J, de Jong P, Catanese J J, Osoegawa K, Shizuya H, Choi S, Chen Y J, Szustakowki J, and International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409(6822):860–921
Leonelli S (2016) Data-centric biology: a philosophical study. University of Chicago Press, Chicago
Lettre G, Jackson A. U., Gieger C., Schumacher F. R., Berndt S. I., Sanna S., Eyheramendy S., Voight B. F., Butler J. L., Guiducci C., Illig T., Hackett R., Heid I. M., Jacobs K. B., Lyssenko V., Uda M., Boehnke M., Chanock S. J., Groop L. C., Hu F. B., Isomaa B., Kraft P., Peltonen L., Salomaa V., Schlessinger D., Hunter D. J., Hayes R. B., Abecasis G. R., Wichmann H.-E., Mohlke K. L., Hirschhorn J. N., Initiative T. D. G., FUSION, KORA, The Prostate, LC, Trial OCS, Study TNH, SardiNIA (2008) Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet 40(5):584–591. https://doi.org/10.1038/ng.125
López-Rubio E, Ratti E (2021) Data science and molecular biology: prediction and mechanistic explanation. Synthese 198(4):3131–3156. https://doi.org/10.1007/s11229-019-02271-0
Matthews LJ, Turkheimer E (2019) Across the great divide: pluralism and the hunt for missing heritability. Synthese. https://doi.org/10.1007/s11229-019-02205-w
Mountjoy E, Schmidt EM, Carmona M, Peat G, Miranda A, Fumis L, Hayhurst J, Buniello A, Schwartzentruber J, Karim MA, Wright D, Hercules A, Papa E, Fauman E, Barrett JC, Todd JA, Ochoa D, Dunham I, Ghoussaini M (2020) Open targets genetics: an open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci, bioRxiv. https://www.biorxiv.org/content/early/2020/09/21/2020.09.16.299271
Needham E, Parker B, Burykin T, James D, Humphreys S (2019) Illuminating the dark phosphoproteome, Sci Signal. Vol. 12
Norton J (1995) Eliminative induction as a method of discovery: how Einstein discovered general relativity. In: Leplin J (ed) The creation of ideas in physics. Kluwer, Alphen aan den Rijn, pp 29–69
O‘Malley M, Elliott K, Burian R (2010) From genetic to genomic regulation: iterativity in microRNA research. Stud Hist Philos Sci Part C Stud Hist Philos Biol Biomed Sci 41(4):407–417
Ratti E (2015) Big Data biology: between eliminative inferences and exploratory experiments. Philos Sci 82:198–218
Ratti E (2020) What kind of novelties can machine learning possibly generate? The case of genomics. Stud Hist Philos Sci Part A. 83:86–96. https://www.sciencedirect.com/science/article/pii/S0039368119302924
Reimers M, Craver C, Dozmorov M, Bacanu S-A, Kendler K (2019) The coherence problem: finding meaning in GWAS complexity. Behav Genet 49:187–195
Richardson S, Stevens H (2015) Postgenomics: perspectives on biology after the genome. Duke University Press, Durham
Samuelson W, Zeckhauser R (1988) Status quo bias in decision making. J Risk Uncertain 1(1):7–59. https://doi.org/10.1007/BF00055564
Schwartzentruber J, Cooper S, Liu JZ, Barrio-Hernandez I, Bello E, Kumasaka N, Young AMH, Franklin RJM, Johnson T, Estrada K, Gaffney DJ, Beltrao P, Bassett A (2021) Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat Genet 53(3):392–402. https://doi.org/10.1038/s41588-020-00776-w
Shalem O, Sanjana NE, Zhang F (2015) High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16(5):299–311
Sharma S, Petsalaki E (2019) Large-scale datasets uncovering cell signalling networks in cancer: context matters. Curr Opin Genet Dev. 54:118–124 Cancer Genomics. https://www.sciencedirect.com/science/article/pii/S0959437X18301278
Steinle F (1997) Entering new fields: exploratory uses of experimentation. Philos Sci. 64:S65–S74. http://www.jstor.org/stable/188390
Sullivan E (2019) Understanding from machine learning models. British J Philos Sci
Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D (2019) Benefits and limitations of genome-wide association studies. Nat Rev Genet 20(8):467–484. https://doi.org/10.1038/s41576-019-0127-1
Weedon MN, Lango H, Lindgren CM, Wallace C, Evans, DM, Mangino M, Freathy RM, Perry J. RB, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann, JS, Vollenweider, P, Waterworth DM, Mooser V, Palmer CNA Morris AD Ouwehand WH, Zhao JH, Li S, Loos R JF, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM, Initiative, DG, Consortium TWTCC, Consortium, CG (2008) Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet 40(5):575–583. https://doi.org/10.1038/ng.121
Wilkes EH, Terfve C, Gribben JG, Saez-Rodriguez J, Cutillas PR (2015) Empirical inference of circuitry and plasticity in a kinase signaling network. Proc Natl Acad Sci U S A 112(25):7719–7724
Wray N, Wijmenga C, Sullivan P, Yang J, Visscher P (2018) Common disease is more complex than implied by the core gene omnigenic model. Cell 173:1573–1580
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42(7):565–569. https://doi.org/10.1038/ng.608
Acknowledgements
We are grateful to Simon Davis, Katie de Lange, and the anonymous reviewers (one of whom turned out to be Pierrick Bourrat) for helpful discussions and feedback. S.S. is supported by a Sir Henry Wellcome Postdoctoral Fellowship at the University of Oxford.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Read, J., Sharma, S. Hypothesis-driven science in large-scale studies: the case of GWAS. Biol Philos 36, 46 (2021). https://doi.org/10.1007/s10539-021-09823-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10539-021-09823-0