Abstract
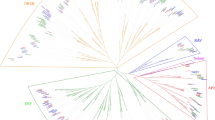
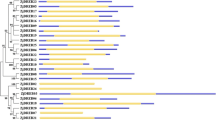
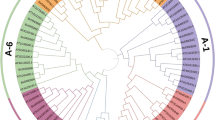
The dehydration responsive element binding (DREB) transcription factor (TF) family comprises unique and important proteins involved in abiotic stress responses and tolerance in plants. Although DREB TFs have been well identified and characterized in a few model plants, there is no detailed information available for mulberry. In this study, 110 AP2/ERF family genes were identified based on a genome-wide analysis of the Morus genome database. Among them, 30 Morus notabilis DREB family genes (MnDREBs) were identified. A comparative analysis with DREB gene families from other plants suggests that MnDREBs could be divided into six subgroups (A-1 to A-6) and could have similar functions in response to abiotic stresses since they have similar conserved domains/motifs within each subgroup. The expression patterns of MnDREBs were analyzed using transcriptome data of different organs from M. notabilis and the quantitative real-time polymerase chain reaction. The expression of most MnDREBs was detected in different organs and induced by various abiotic stresses, which suggest their vital roles in abiotic stress tolerance.
Similar content being viewed by others
Abbreviations
- ABRE:
-
element involved in the abscisic acid responsiveness
- AP2:
-
APETALA2
- ARE:
-
element for the anaerobic induction
- CBF1:
-
CRT/DRE-binding factor 1
- CDS:
-
corresponding coding sequence
- DRE:
-
dehydration-responsive element
- DRTF:
-
database of rice transcription factors
- DREB:
-
dehydration responsive element binding
- ERE:
-
ethylene-responsive element
- ERF:
-
ethylene responsive element binding factors
- GARE:
-
gibberellin-responsive element
- Glu:
-
glutamine
- GWD:
-
genome-wide duplications
- HSE:
-
element involved in heat stress responsiveness
- LTR:
-
element involved in low-temperature responsiveness
- MBS:
-
MYB binding site
- MBSI:MYB:
-
binding site involved in flavonoid biosynthetic genes regulation
- NCBI:
-
National Center for Biotechnology Information
- NJ:
-
neighbor-joining
- NLS:
-
nuclear localization sequence
- ORF:
-
open reading frame
- pI:
-
isoelectric point
- plant TFDB:
-
plant transcription factor database
- RAV:
-
related to ABI3/VP1
- RPL15:
-
ribosomal protein L15
- RT-qPCR:
-
real-time quantitative polymerase chain reaction
- SMART:
-
simple modular architecture research tool
- TAIR:
-
Arabidopsis information resource
- TF:
-
transcription factor
- Val:
-
valine
- WUN:
-
wound-responsive element
References
Agarwal, P., Agarwal, P.K., Nair, S., Sopory, S.K., Reddy, M.K.: Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity. — Mol. Genet. Genomics 277: 189–198, 2007.
Agarwal, P.K., Agarwal, P., Reddy, M.K., Sopory, S.K.: Role of DREB transcription factors in abiotic and biotic stress tolerance in plant. — Plant Cell Rep. 25: 1263–1274, 2006.
Agarwal, P.K., Jha, B.: Transcription factors in plants and ABA dependent and independent abiotic stress signaling. — Biol. Plant. 54: 201–212, 2010.
Akhtar, M., Jaiswal, A., Taj, G., Jaiswal, J.P., Qureshi, M.I., Singh, N.K.: DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. — J. Genet. 91: 385–395, 2012.
Bouaziz, D., Pirrello, J., Ben Amor, H., Hammami, A., Charfeddine, M., Dhieb, A., Bouzayen, M., Gargouri-Bouzid, R.: Ectopic expression of dehydration responsive element binding proteins (StDREB2) confers higher tolerance to salt stress in potato. — Plant Physiol. Biochem. 60: 98–108, 2012.
Boyer, J.S.: Plant Productivity and Environment. — Science 218: 443–448, 1982.
Checker, V.G., Khurana, P.: Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. — Plant Cell Rep. 32: 1729–1741, 2013.
Chen, M., Wang, Q.Y., Cheng, X.G., Xu, Z.S., Li, L.C., Ye, X.G., Xia, L.Q., Ma, Y.Z.: GmDREB2, a soybean DREbinding transcription factor, conferred drought and high-salt tolerance in transgenic plants. — Biochem. biophys. Res. Commun. 353: 299–305, 2007.
Chen, Y., Yang. J., Wang, Z., Zhang, H., Mao, X., Li, C.: Gene structures, classification, and expression models of the DREB transcription factor subfamily in Populus trichocarpa. — Sci. World J. doi.org/10.1155/2013/954640, 2013.
Cui, M., Zhang, W., Zhang, Q., Xu, Z., Zhu, Z., Duan, F., Wu, R.: Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. — Plant Physiol. Biochem. 49: 1384–1391, 2011.
Dong, C.J., Liu, J.Y.: The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. — BMC Plant Biol. 10: 47, 2010.
Dubouzet, J.G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E.G., Miura, S., Seki, M., Shinozaki, K., Yamaguchi-Shinozaki, K.: OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought, high salt and cold responsive gene expression. — Plant J. 33: 751–763, 2003.
Egawa, C., Kobayashi, F., Ishibashi M., Nakamura, T., Nakamura, C., Takumi, S.: Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. — Genes. Genet. Syst. 81: 77–91, 2006.
Guo, A.Y., Zhu, Q.H., Chen, X., Luo, J.C.: GSDS: a gene structure display server. — Yi Chuan 29: 1023–1026, 2007.
Hasegawa, P.M., Bressan, R.A., Zhu, J.K., Bohnert, H.J.: Plant cellular and molecular responses to high salinity. — Annu. Rev. Plant Physiol. Plant mol. Biol. 51: 463–499, 2000.
He, N., Zhang, C., Qi, X., Zhao, S., Tao, Y., Yang, G., Lee, T.H., Wang, X., Cai, L., Li, D., Lu, M., Liao, S., Luo, G., He, R., Tan, H., Xu, Y., Li, T., Zhao, A., Jia, L., Fu, Q., Zeng, Q., Gao, C., Ma, B., Liang, J., Wang, X., Shang, J., Song, P., Wu, H., Fan, L., Wang, Q., Shuai, Q., Zhu, J., Wei, C., Zhu-Salzman, K., Jin, D., Wang, J., Liu, T., Yu, M., Tang, C., Wang, Z., Dai, F., Chen, J., Liu, Y., Zhao, S., Lin, T., Zhang, S., Wang, J., Wang, J., Yang, H., Yang, G., Wang, J., Paterson, A.H., Xia, Q., Ji, D.: Draft genome sequence of the mulberry tree Morus notabilis. — Nat. Commun. 4: 2445, 2013.
Huang, G.T., Ma, S.L., Bai, L.P., Zhang, L., Ma, H., Jia, P., Liu, J., Zhong, M., Guo, Z.F.: Signal transduction during cold, salt, and drought stresses in plants. — Mol. Biol. Rep. 39: 969–987, 2012.
Hussain, S.S., Kayani, M.A., Amjad, M.: Transcription factors as tools to engineer enhanced drought stress tolerance in plants. — Biotechnol. Progr. 27: 297–306, 2011.
Hwang, J.E., Lim, C.J., Chen, H., Je, J., Song, C., Lim, C.O.: Overexpression of Arabidopsis dehydration-responsive element-binding protein 2C confers tolerance to oxidative stress. — Mol. Cells 33: 135–140, 2012.
Islam, M.S., Wang, M.H.: Expression of dehydration responsive element-binding protein-3 (DREB3) under different abiotic stresses in tomato. — BMB Rep. 42: 611–616, 2009.
Jaillon, O., Aury, J.M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., Choisne, N., Aubourg, S., Vitulo, N., Jubin, C., Vezzi, A., Legeai, F., Hugueney, P., Dasilva, C., Horner, D., Mica, E., Jublot, D., Poulain, J., Bruyère, C., Billault, A., Segurens, B., Gouyvenoux, M., Ugarte, E., Cattonaro, F., Anthouard, V., Vico, V., Fabbro, C.D., Alaux, M., Gaspero, G.D., Dumas, V., Felice, N., Paillard, S., Juman, I., Moroldo, M., Scalabrin, S., Canaguier, A., Clainche, I.L., Malacrida, G., Durand, E., Pesole, G., Laucou, V., Chatelet, P., Merdinoglu, D., Delledonne, M., Pezzotti, M., Lecharny, A., Scarpelli, C., Artiguenave, F., Pè, M.E., Valle, G., Morgante, M., Caboche, M., Adam-Blondon, A.F., Weissenbach, J., Quétier, F., Wincker, P.: The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. — Nature 449: 463–467, 2007.
Jofuku, K.D., Den Boer, B.G., Van Montagu, M., Okamuro, J.K.: Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. — Plant Cell 6: 1211–1225, 1994.
Kizis, D., Pages, M.: Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought responsive element in an ABA-dependent pathway. — Plant J. 30: 679–689, 2002.
Lata, C., Prasad, M.: Role of DREBs in regulation of abiotic stress responses in plants. — J. exp. Bot. 62: 4731–4748, 2011.
Lee, S.J., Kang, J.Y., Park, H.J., Kim, H.D., Bae, M.S., Choi, Choi, H.I., Kim, S.Y.: DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsivegene expression, and its overexpression affects abscisic acid sensitivity. — Plant Physiol. 153: 716–727, 2010.
Li, M., Li, Y., Li, H., Wu, G.: Improvement of paper mulberry tolerance to abiotic stresses by ectopic expression of tall fescue FaDREB1. — Tree Physiol. 32: 104–113, 2011a.
Li, M.R., Li, Y., Li, H.Q., Wu, G.J.: Ectopic expression of FaDREB2 enhances osmotic tolerance in paper mulberry. — J. Integr. Plant Biol. 53: 951–960, 2011b.
Lucas, S., Durmaz, E., Akpınar, B.A., Budak, H.: The drought response displayed by a DRE-binding protein from Triticum dicoccoides. — Plant Physiol. Biochem. 49, 346–351, 2011.
Marcolino-Gomes, J., Rodrigues, F.A., Oliveira, M.C., Farias, J.R., Neumaier, N., Abdelnoor, R.V., Marcelino-Guimarães, F.C., Nepomuceno, A.L.: Expression patterns of GmAP2/EREB-like transcription factors involved in soybean responses to water deficit. — PLOS ONE. 8: e62294, 2013.
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., Shinozaki, K., Yamaguchi-Shinozaki, K.: Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. — Mol. Genet. Genomics 283: 185–196, 2010.
Mayer, K., White, O., Bevan, M., Lemcke, K., Creasy, T.H., Bielke, C., Haas, B., Haase, D., Maiti, R., Rudd, S., Peterson, J., Schoof, H., Frishman, D., Morgenstern, B., Zaccaria, P., Ermolaeva, M., Pertea, M., Quackenbush, J., Volfovsky, N., Wu, D., Lowe, T.M., Salzberg, S.L., Mewes, H.W.: Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. — Nature 408: 796–815, 2000.
Nakano, T., Suzuki, K., Fujimura, T., Shinshi, H.: Genomewide analysis of the ERF gene family in Arabidopsis and rice. — Plant Physiol. 140: 411–432, 2006.
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L.S., Shinozaki, K., Yamaguchi-Shinozaki, K.: Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. — Plant J. 50: 54–69, 2007.
Ramachandra Reddy, A., Chaitanya, K.V., Jutur, P.P., Sumithra, K.: Differential antioxidative responses to water stress among five mulberry (Morus alba L.) cultivars. — Environ. exp. Bot. 52: 33–42, 2004.
Saitou, N., Nei, M.: The neighbor-joining method: a new method for reconstructing phylogenetic trees. — Mol. Biol. Evol. 4: 406–425, 1987.
Sakuma, Y., Liu, Q., Dubouzet, J.G., Abe, H., Shinozaki, K., Yamaguchi-Shinozaki, K.: DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydrationand cold-inducible gene expression. — Biochem. biophys. Res. Commun. 290: 998–1009, 2002.
Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., Shinozaki, K.: Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. — Plant Cell 13: 61–72, 2001.
Sharma, M.K., Kumar, R., Solanke, A.U., Sharma, R., Tyagi, A.K., Sharma, A.K.: Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. — Mol. Genet. Genomics 284: 455–475, 2010.
Sharoni, A.M.1., Nuruzzaman, M., Satoh, K., Shimizu, T., Kondoh, H., Sasaya, T., Choi, I.R., Omura, T., Kikuchi, S.: Gene structures, classification and expression models of the AP2/ERFBP transcripttion factor family in rice. — Plant Cell Physiol. 52: 344–360, 2011.
Shen, Y.G., Zhang, W.K., He, S.J., Zhang, J.S., Liu, Q., Chen, S.Y.: An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. — Theor. appl. Genet. 106: 923–930, 2003.
Singhal, B.K., Khan, M.A., Dhar, A., Baqual, F.M., Bindroo, B.B.: Approaches to industrial exploitation of mulberry (Mulberry sp.) fruits. — J. Fruit Ornam. Plant Res. 18: 83–99, 2010.
Stockinger, E.J., Gilmour, S.J., Thomashow, M.F.: Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. — Proc. nat. Acad. Sci. USA 94: 1035–104, 1997.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S.: MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. — Mol. Biol. Evol. 28: 2731–2739, 2011.
Tang, M., Liu, X., Deng, H., Shen, S.: Over-expression of JcDREB, a putative AP2/EREBP domain-containing transcription factor gene in woody biodiesel plant Jatropha curcas, enhances salt and freezing tolerance in transgenic Arabidopsis thaliana. — Plant Sci. 181: 623–631, 2011.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G.: The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. — Nucl. Acids Res. 25: 4876–4882, 1997.
Tuskan, G.A., Difazio, S., Jansson, S., Bohlmann, J., Grigorive, I., Hellsten, U., Putnam, N., Ralph, S., Rombauts, S., Salamov, A., Schein, J., Sterck, L., Aerts, A., Bhalerao, R. R., Bhalerao, R. P., Blaudez, D., Boerjan, W., Brun, A., Brunner, A., Busov, V., Campbell, M., Carlson, J., Chalot, M., Chapman, J., Chen, G.L., Cooper, D., Coutinho, P. M., Couturier, J., Covert, S., Cronk, Q., Cunningham, R., Davis, J., Degroeve, S., Déjardin, A., De Pamphilis, C., Detter, J., Dirks, B., Dubchak, I., Duplessis, S., Ehlting, J., Ellis, B., Gendler, K., Goodstein, D., Gribskov, M., Grimwood, J., Groover, A., Gunter, L., Hamberger, B., Heinze, B., Helariutta, Y., Henrissat, B., Holligan, D., Holt, R., Huang, W., Islam-Faridi, N., Jones, S., Jones-Rhoades, M., Jorgensen, R., Joshi, C., Kangasjärvi, J., Karlsson, J., Kelleher, C., Kirkpatrick, R., Kirst, M., Kohler, A., Kalluri, U., Larimer, F., Leebens-Mack, J., Leplé, J.C., Locascio, P., Lou, Y., Lucas, S., Martin, F., Montanini, B., Napoli, C., Nelson, D. R., Nelson, C., Nieminen, K., Nilsson, O., Pereda, V., Peter, G., Philippe, R., Pilate, G., Poliakov, A., Razumovskaya, J., Richardson, P., Rinaldi, C., Ritland, K., Rouzé, P., Ryaboy, D., Schmutz, J., Schrader, J., Segerman, B., Shin, H., Siddiqui, A., Sterky, F., Terry, A., Tsai, C.J., Uberbacher, E., Unneberg, P., Vahala, J., Wall, K., Wessler, S., Yang, G., Yin, T., Douglas, C., Marra, M., Sandberg, G., Van de Peer, Y., Rokhsar, D.: The Genome of black cottonwood, Populus trichocarpa (Torr. & Gray). — Science 313: 1596–1604, 2006.
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., Fontana, P., Bhatnagar, S.K., Troggio, M., Pruss, D., Salvi, S., Pindo, M., Baldi, P., Castelletti, S., Cavaiuolo, M., Coppola, G., Costa, F., Cova, V., Ri, A.D., Goremykin, V., Komjanc, M., Longhi, S., Magnago, P., Malacarne, G., Malnoy, M., Micheletti, D., Moretto, M., Perazzolli, M., Si-Ammour, A., Vezzulli, S., Zini, E., Eldredge, G., Fitzgerald, L.M., Gutin, N., Lanchbury, J., Macalma, T., Mitchell, J.T., Reid, J., Wardell, B., Kodira, C., Chen, Z., Desany, B., Niazi, F., Palmer, M., Koepke, T., Jiwan, D., Schaeffer, S., Krishnan, V., Wu, C., Chu, V.T., King, S.T., Vick, J., Tao, Q., Mraz, A., Stormo, A., Stormo, K., Bogden, R., Ederle, D., Stella, A., Vecchietti, A., Kater, M.M., Masiero, S., Lasserre, P., Lespinasse, Y., Allan, A.C., Bus, V., Chagné, D., Crowhurst, R.N., Gleave, A.P., Lavezzo, E., Fawcett, J.A., Proost, S., Rouzé, P., Sterck, L., Toppo, S., Lazzari, B., Hellens, R.P., Durel, C.E., Gutin, A., Bumgarner, R.E., Gardiner, S.E., Skolnick, M., Egholm, M., Van de Peer, Y., Salamini, F., Viola, R.: The genome of the domesticated apple (Malus × domestica Borkh.). — Nat. Genet. 42: 833–839, 2010.
Wei, C., Liu, X., Long, D., Guo, Q., Fang, Y., Bian, C., Zhang, D., Zeng, Q., Xiang, Z., Zhao, A.: Molecular cloning and expression analysis of mulberry MAPK gene family. — Plant Physiol. Biochem. 77C: 108–116, 2014.
Xue, G.P., Loveridge, C.W.: HvDRF1 is involved in abscisic acid mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. — Plant J. 37: 326–339, 2004.
Xue, Y., Wang, Y.Y., Peng, R.H., Zhen, J.L., Zhu, B., Gao, J.J., Zhao, W., Han, H.J., Yao, Q.H.: Transcription factor MdCBF1 gene increases freezing stress tolerance in transgenic Arabidopsis thaliana. — Biol. Plant. 58: 499–506, 2014.
Yu, J., Hu, S., Wang, J., Wong, G.K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., Cao, M., Liu, J., Sun, J., Tang, J., Chen, Y., Huang, X., Lin, W., Ye, C., Tong, W., Cong, L., Geng, J., Han, Y., Li, L., Li, W., Hu, G., Huang, X., Li, W., Li, J., Liu, Z., Li, L., Liu, J., Qi, Q., Liu, J., Li, L., Li, T., Wang, X., Lu, H., Wu, T., Zhu, M., Ni, P., Han, H., Dong, W., Ren, X., Feng, X., Cui, P., Li, X., Wang, H., Xu, X., Zhai, W., Xu, Z., Zhang, J., He, S., Zhang, J., Xu, J., Zhang, K., Zheng, X., Dong, J., Zeng, W., Tao, L., Ye, J., Tan, J., Ren, X., Chen, X., He, J., Liu, D., Tian, W., Tian, C., Xia, H., Bao, Q., Li, G., Gao, H., Cao, T., Wang, J., Zhao, W., Li, P., Chen, W., Wang, X., Zhang, Y., Hu, J., Wang, J., Liu, S., Yang, J., Zhang, G., Xiong, Y., Li, Z., Mao, L., Zhou, C., Zhu, Z., Chen, R., Hao, B., Zheng, W., Chen, S., Guo, W., Li, G., Liu, S., Tao, M., Wang, J., Zhu, L., Yuan, L., Yang, H.: A draft sequence of the rice genome (Oryza sativa L. ssp.indica). — Science 296: 79–92, 2002.
Zhao, T., Liang, D., Wang, P., Liu, J., Ma, F.: Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. — Mol. Genet. Genomics 287: 423–436, 2012.
Zhao, T., Xia, H., Liu, G., Ma, F.: The gene family of dehydration responsive element-binding transcription factors in grape (Vitis vinifera): genome-wide identification and analysis, expression profiles, and involvement in abiotic stress resistance. — Mol. Biol. Rep. 41: 1577–1590, 2014.
Zhou, M.L., Ma, J.T., Zhao, Y.M., Wei, Y.H., Tang, Y.X., Wu, Y.M.: Improvement of drought and salt tolerance in Arabidopsis and Lotus corniculatus by overexpression of a novel DREB transcription factor from Populus euphratica. — Gene 506: 10–17, 2012.
Zhu, T., Budworth, P., Han, B., Brown, D., Chang, H.S., Zou, G.Z., Wang, X.: Toward elucidating the global expression patterns of developing Arabidopsis: parallel analysis of 8 300 genes by a high-density oligonucleotide probe array. — Plant Physiol. Biochem. 39: 221–242, 2001.
Zhuang, J., Cai, B., Peng, R.H., Zhu, B., Jin, X.F., Xue, Y., Gao, F., Fu, X.Y., Tian, Y.S., Zhao, W., Qiao, Y.S., Zhang, Z., Xiong, A.S., Yao, Q.H.: Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. — Biochem. biophys. Res. Commun. 371: 468–474, 2008.
Zhuang, J., Peng, R.H., Cheng, Z.M., Zhang, J., Cai, B., Zhang, Z., Gao, F., Zhu, B., Fu, X.Y., Jin, X.F., Chen, J.M., Qiao, Y.S., Xiong, A.S., Yao, Q.H.: Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. — Scientia Hort. 123: 73–81, 2009.
Author information
Authors and Affiliations
Corresponding author
Additional information
Acknowledgements: This work was supported by grants from the Chongqing Science & Technology Commission (No. cstc2012jjA80039), the China Agriculture Research System (No. CARS-22-ZJ0102), and the National Hi-Tech Research and Development Program of China (No. 2013AA100605-3). The first two authors contributed equally to this paper.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Liu, X.Q., Zhu, J.J., Wei, C.J. et al. Genome-wide identification and characterization of the DREB transcription factor gene family in mulberry. Biol Plant 59, 253–265 (2015). https://doi.org/10.1007/s10535-015-0498-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10535-015-0498-x