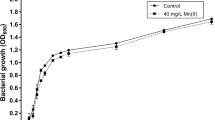
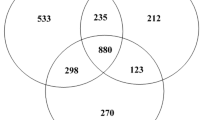
Abstract
Bacillus toyonensis SFC 500-1E is a member of the consortium SFC 500-1 able to remove Cr(VI) and simultaneously tolerate high phenol concentrations. In order to elucidate mechanisms utilized by this strain during the bioremediation process, the differential expression pattern of proteins was analyzed when it grew with or without Cr(VI) (10 mg/L) and Cr(VI) + phenol (10 and 300 mg/L), through two complementary proteomic approaches: gel-based (Gel-LC) and gel-free (shotgun) nanoUHPLC-ESI–MS/MS. A total of 400 differentially expressed proteins were identified, out of which 152 proteins were down-regulated under Cr(VI) and 205 up-regulated in the presence of Cr(VI) + phenol, suggesting the extra effort made by the strain to adapt itself and keep growing when phenol was also added. The major metabolic pathways affected include carbohydrate and energetic metabolism, followed by lipid and amino acid metabolism. Particularly interesting were also ABC transporters and the iron-siderophore transporter as well as transcriptional regulators that can bind metals. Stress-associated global response involving the expression of thioredoxins, SOS response, and chaperones appears to be crucial for the survival of this strain under treatment with both contaminants. This research not only provided a deeper understanding of B. toyonensis SFC 500-1E metabolic role in Cr(VI) and phenol bioremediation process but also allowed us to complete an overview of the consortium SFC 500-1 behavior. This may contribute to an improvement in its use as a bioremediation strategy and also provides a baseline for further research.
Graphical abstract






Similar content being viewed by others
References
Aebersold R, Mann M (2016) Mass-spectrometric exploration of proteome structure and function. Nature 537:347–355
Ahemad M (2014) Bacterial mechanisms for Cr(VI) resistance and reduction: an overview and recent advances. Folia Microbiol (Praha) 59:321–332
Antelmann H, Hecker M, Zuber P (2008) Proteomic signatures uncover thiol-specific electrophile resistance mechanisms in Bacillus subtilis. Expert Rev Proteomics 5:77–90
APHA-AWWA (1989) Standard methods for the examination of water and wastewater, 17th Ed. American Public Health Association, Washington DC
Bae WC, Lee HK, Choe YC et al (2005) Purification and characterization of NADPH-dependent Cr(VI) reductase from Escherichia coli ATCC 33456. J Microbiol 43:21–27
Baksh KA, Zamble DB (2020) Allosteric control of metal-responsive transcriptional regulators in bacteria. J Biol Chem 295:1673–1684
Bansal N, Coetzee JJ, Chirwa EMN (2019) In situ bioremediation of hexavalent chromium in presence of iron by dried sludge bacteria exposed to high chromium concentration. Ecotoxicol Environ Saf 172:281–289
Bellenberg S, Huynh D, Poetsch A et al (2019) Proteomics reveal enhanced oxidative stress responses and metabolic adaptation in Acidithiobacillus ferrooxidans biofilm cells on pyrite. Front Microbiol 10:1–14
Berg K, Pedersen HL, Leiros I (2020) Biochemical characterization of ferric uptake regulator (Fur) from Aliivibrio salmonicida. mapping the DNA sequence specificity through binding studies and structural modelling. Biometals 33:169–185
Beringer JE (1974) R factor transfer in Rhizobiurn zegurninosarum. J Gen Microbiol 84:188–198
Bhat SV, Booth SC, Vantomme EAN et al (2015) Oxidative stress and metabolic perturbations in Escherichia coli exposed to sublethal levels of 2,4-dichlorophenoxyacetic acid. Chemosphere 135:453–461
Bhattacharya P, Barnebey A, Zemla M et al (2015) Complete genome sequence of the chromate-reducing bacterium Thermoanaerobacter thermohydrosulfuricus strain BSB-33. Stand Genomic Sci 10:1–13
Bonilla JO, Callegari EA, Delfini CD et al (2016) Simultaneous chromate and sulfate removal by Streptomyces sp. MC1. changes in intracellular protein profile induced by Cr(VI). J Basic Microbiol 56:1212–1221
Bonilla JO, Callegari EA, Estevéz MC, Villegas LB (2020) Intracellular proteomic analysis of Streptomyces sp. MC1 when exposed to Cr(VI) by gel-based and gel-free methods. Curr Microbiol 77:62–70
Bonilla JO, Callegari EA, Paez MD et al (2021) Characterization of copper stress response in Fusarium tricinctum M6: a metal-resistant microorganism isolated from an acid mine drainage-affected environment. J Hazard Mater 412:125216
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 254:248–254
Brown SD, Thompson MR, VerBerkmoes NC et al (2006) Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol Cell Proteomics 5:1054–1071
Butcher J, Sarvan S, Brunzelle JS et al (2012) Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci USA 109:10047–10052
Callegari EA (2016) Shotgun proteomics analysis of estrogen effects in the uterus using two-dimensional liquid chromatography and tandem mass spectrometry. In: Eyster KM (ed) Estrogen receptors: methods and protocols. Springer, New York, pp 131–148
Ceylan S, Akbulut BS, Denizci AA, Kazan D (2011) Proteomic insight into phenolic adaptation of a moderately halophilic Halomonas sp. strain AAD12. Can J Microbiol 57:295–302. https://doi.org/10.1139/w11-009
Chen C, Hou J, Tanner JJ, Cheng J (2020) Bioinformatics methods for mass spectrometry-based proteomics data analysis. Int J Mol Sci 21:2873
Cheng B, Li S, Wang M, Li Y (2015) Investigation of combined pollution between mand a variety of pollutants based on the fractional factorial design. Pol J Environ Stud 24:1939–1947. https://doi.org/10.15244/pjoes/42354
Chourey K, Thompson MR, Morrell-Falvey J et al (2006) Global molecular and morphological effects of 24-hour chromium(VI) exposure on Shewanella oneidensis MR-1. Appl Environ Microbiol 72:6331–6344
Dawson LF, Valiente E, Faulds-Pain A et al (2012) Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. PLoS ONE 7:1–13
Decorosi F (2010) Studio di ceppi batterici per il biorisanamento di suoli contaminati da Cr(VI). Firenze University Press, Florence
Decorosi F, Lori L, Santopolo L et al (2011) Characterization of a Cr(VI)-sensitive Pseudomonas corrugata 28 mutant impaired in a pyridine nucleotide transhydrogenase gene. Res Microbiol 162:747–755
Deghles A, Kurt U (2016) Treatment of raw tannery wastewater by electrocoagulation technique: optimization of effective parameters using Taguchi method. Desalin Water Treat 57:14798–14809
Dekker L, Arsène-Ploetze F, Santini JM (2016) Comparative proteomics of Acidithiobacillus ferrooxidans grown in the presence and absence of uranium. Res Microbiol 167:234–239
Deng W, Li C, Xie J (2013) The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell Signal 25:1608–1613
Dong F-X, Yan L, Zhou X-H et al (2021) Simultaneous adsorption of Cr(VI) and phenol by biochar-based iron oxide composites in water: performance, kinetics and mechanism. J Hazard Mater 416:125930
Fernandez M, Paisio CE, Perotti R et al (2019a) Laboratory and field microcosms as useful experimental systems to study the bioaugmentation treatment of tannery effluents. J Environ Manag 234:503–511
Fernandez M, Paulucci NS, Peppino Margutti M et al (2019b) Membrane rigidity and phosphatidic acid (PtdOH) signal: two important events in Acinetobacter guillouiae SFC 500-1A exposed to chromium(VI) and phenol. Lipids 54:557–570
Fernandez M, Morales GM, Agostini E, González PS (2020) Morphological and structural response of Bacillus sp. SFC 500-1E after Cr(VI) and phenol treatment. J Basic Microbiol 60:679–690
Gang H, Xiao C, Xiao Y et al (2019) Proteomic analysis of the reduction and resistance mechanisms of Shewanella oneidensis MR-1 under long-term hexavalent chromium stress. Environ Int 127:94–102
Gao L, Zhou Z, Chen X et al (2020) Comparative proteomics analysis reveals new features of the oxidative stress response in the polyextremophilic bacterium Deinococcus radiodurans. Microorganisms 8:451
Garmory HS, Titball RW (2004) ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72:6757–6763
Gonzalez CF, Ackerley DF, Lynch SV, Matin A (2005) ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J Biol Chem 280:22590–5
Gu Q, Wu Q, Zhang J et al (2017) Acinetobacter sp. DW-1 immobilized on polyhedron hollow polypropylene balls and analysis of transcriptome and proteome of the bacterium during phenol biodegradation process. Sci Rep 7:4863
Gunasekera TS, Csonka LN, Paliy O (2008) Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol 190:3712–3720
Guo H, Wang L, Deng Y, Ye J (2021) Novel perspectives of environmental proteomics. Sci Total Environ 788:147588
Haggett L, Bhasin A, Srivastava P, Fujita M (2018) A revised model for the control of fatty acid synthesis by master regulator Spo0A in Bacillus subtilis. Mol Microbiol 108:424–442
Henne KL, Turse JE, Nicora CD et al (2009) Global proteomic analysis of the chromate response in Arthrobacter sp. strain FB24. J Proteome Res 8:1704–1716
Heyer R, Kohrs F, Reichl U, Benndorf D (2015) Metaproteomics of complex microbial communities in biogas plants. Microb Biotechnol 8:749–763
Heyer R, Schallert K, Zoun R et al (2017) Challenges and perspectives of metaproteomic data analysis. J Biotechnol 261:24–36
Hu X, Boyer GL (1996) Siderophore-mediated aluminum uptake by Bacillus megaterium ATCC 19213. Appl Environ Microbiol 62:4044–4048
Huynh PL, Jankovic I, Schnell NF, Brückner R (2000) Characterization of an HPr kinase mutant of Staphylococcus xylosus. J Bacteriol 182:1895–1902
Izrael-Živkovic L, Rikalović M, Gojgić-Cvijović G et al (2018) Cadmium specific proteomic responses of a highly resistant: Pseudomonas aeruginosa san ai. RSC Adv 8:10549–10560
Jiang K, Zhang J, Deng Z et al (2021) Natural attenuation mechanism of hexavalent chromium in a wetland: zoning characteristics of abiotic and biotic effects. Environ Pollut 287:117639
Joutey NT, Sayel H, Bahafid W, Ghachtouli E (2015) Mechanisms of hexavalent chromium resistance and removal by microorganisms. Rev Environ Contam Toxicol 233:45–70
Kanehisa M, Sato Y, Morishima K (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731
Kelstrup CD, Young C, Lavallee R et al (2012) Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J Proteome Res 11:3487–3497
Kim H, Wu K, Lee C (2021) Stress-responsive periplasmic chaperones in bacteria. Front Mol Biosci 8:1–14
Koçberber N, Stensballe A, Erik D, Dönmez G (2010) Proteomic changes in response to chromium(VI) toxicity in Pseudomonas aeruginosa. Bioresour Technol 101:2134–2140
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Li F, Song W, Wei J et al (2016) Comparative proteomic analysis of phenol degradation process by Arthrobacter. Int Biodeterior Biodegrad 110:189–198
Liu Y, Min J-W, Feng S et al (2020) Therapeutic role of a cysteine precursor, OTC, in ischemic stroke is mediated by improved proteostasis in mice. Transl Stroke Res 11:147–160
Liu H, Zhang Y, Wang Y et al (2021) The connection between Czc and Cad systems involved in cadmium resistance in Pseudomonas putida. Int J Mol Sci 22:1–14
López-Pedrouso M, Varela Z, Franco D et al (2020) Can proteomics contribute to biomonitoring of aquatic pollution? A critical review. Environ Pollut 267:115473
Ma Z, Jacobsen FE, Giedroc DP (2009) Coordination chemistry of bacterial metal transport and sensing. Chem Rev 109:4644–4681
Ma L, Xu J, Chen N et al (2019) Microbial reduction fate of chromium (Cr) in aqueous solution by mixed bacterial consortium. Ecotoxicol Environ Saf 170:763–770
Makarova KS, Aravind L, Wolf YI et al (2001) Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev 65:44–79
Malaviya P, Singh A (2014) Bioremediation of chromium solutions and chromium containing wastewaters. Crit Rev Microbiol 00:1–27
Malherbe G, Humphreys DP, Davé E (2019) A robust fractionation method for protein subcellular localization studies in Escherichia coli. Biotechniques 66:171–178
Malla MA, Dubey A, Yadav S et al (2018) Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front Microbiol 9:1–18
Mandal SK, Adhikari R, Sharma A et al (2019) Designating ligand specificities to metal uptake ABC transporters in Thermus thermophilus HB8. Metallomics 11:597–612
Mastroleo F, Leroy B, Van Houdt R et al (2009) Shotgun proteome analysis of Rhodospirillum rubrum S1H: integrating data from gel-free and gel-based peptides fractionation methods. J Proteome Res 8:2530–2541
Matilda CS, Mannully ST, Viditha RP, Shanthi C (2019) Protein profiling of metal-resistant Bacillus cereus VITSH1. J Appl Microbiol 127:121–133
McConnell RE, Benesh AE, Mao S et al (2011) Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol 300:G914–G926
Meyer TS, Lamberts BL (1965) Use of coomassie brilliant blue R250 for the electrophoresis of microgram quantities of parotid saliva proteins on acrylamide-gel strips. Biochim Biophys Acta 107:144–145
Monoyios A, Hummel K, Nöbauer K et al (2018) An alliance of gel-based and gel-free proteomic techniques displays substantial insight into the proteome of a virulent and an attenuated Histomonas meleagridis strain. Front Cell Infect Microbiol 8:407
Monsieurs P, Moors H, Van Houdt R et al (2011) Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. Biometals 24:1133–1151
Murínová S, Dercová K (2014) Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int J Microbiol 2014:873081
Nilo-Poyanco R, Moraga C, Benedetto G et al (2021) Shotgun proteomics of peach fruit reveals major metabolic pathways associated to ripening. BMC Genomics 22:17
Ontañon OM, González PS, Agostini E (2015a) Biochemical and molecular mechanisms involved in simultaneous phenol and Cr(VI) removal by Acinetobacter guillouiae SFC 500-1A. Environ Sci Pollut Res 22:13014–13023. https://doi.org/10.1007/s11356-015-4571-y
Ontañon OM, González PS, Agostini E (2015b) Optimization of simultaneous removal of Cr(VI) and phenol by a native bacterial consortium: its use for bioaugmentation of co-polluted effluents. J Appl Microbiol 119:1011–1022. https://doi.org/10.1111/jam.12913
Ontañon OM, González PS, Barros GG, Agostini E (2017) Improvement of simultaneous Cr(VI) and phenol removal by an immobilised bacterial consortium and characterisation of biodegradation products. N Biotechnol 37:172–179. https://doi.org/10.1016/j.nbt.2017.02.003
Ontañon OM, Fernandez M, Agostini E, González PS (2018a) Identification of the main mechanisms involved in the tolerance and bioremediation of Cr(VI) by Bacillus sp. SFC 500-1E. Env Sci Pollut Res 25:16111–16120. https://doi.org/10.1007/s11356-018-1764-1
Ontañon OM, Landi C, Carleo A et al (2018b) What makes A. guillouiae SFC 500-1A able to co-metabolize phenol and Cr(VI)? A proteomic approach. J Hazard Mater 354:215–224. https://doi.org/10.1016/j.jhazmat.2018.04.068
Osman D, Cavet JS (2010) Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat Prod Rep 27:668–680. https://doi.org/10.1039/b906682a
Paez MD, Callegari EA (2022) Proteomics analysis of the estrogen effects in the rat uterus using gel-LC and tandem mass spectrometry approaches. In: Eyster KM (ed) Estrogen receptors: methods and protocols. Springer US, New York, pp 289–311
Pagès D, Sanchez L, Conrod S et al (2007) Exploration of intraclonal adaptation mechanisms of Pseudomonas brassicacearum facing cadmium toxicity. Environ Microbiol 9:2820–2835
Pereira EJ, Damare S, Furtado B, Ramaiah N (2018) Response to chromate challenge by marine Staphylococcus sp. NIOMR8 evaluated by differential protein expression. 3 Biotech 8:500. https://doi.org/10.1007/s13205-018-1522-6
Pereira PP, Fernandez M, Cimadoro J et al (2021) Biohybrid membranes for effective bacterial vehiculation and simultaneous removal of hexavalent chromium (CrVI) and phenol. Appl Microbiol Biotechnol 105:827–838
Piersma SR, Warmoes MO, de Wit M et al (2013) Whole gel processing procedure for GeLC-MS/MS based proteomics. Proteome Sci 11:17
Rodionova IA, Zhang Z, Mehla J et al (2017) The phosphocarrier protein HPr of the bacterial phosphotransferase system globally regulates energy metabolism by directly interacting with multiple enzymes in Escherichia coli. J Biol Chem 292:14250–14257
Rodriguez Ayala F, Bartolini M, Grau R (2020) The stress-responsive alternative sigma factor SigB of Bacillus subtilis and its relatives: an old friend with new functions. Front Microbiol 11:1761
Roma-Rodrigues C, Santos PM, Benndorf D et al (2010) Response of Pseudomonas putida KT2440 to phenol at the level of membrane proteome. J Proteomics 73:1461–1478
Saha RP, Samanta S, Patra S et al (2017) Metal homeostasis in bacteria: the role of ArsR–SmtB family of transcriptional repressors in combating varying metal concentrations in the environment. Biometals 30:459–503
Santos T, Viala D, Chambon C et al (2019) Listeria monocytogenes biofilm adaptation to different temperatures seen through shotgun proteomics. Front Nutr 6:1–16
Schalk IJ, Hannauer M, Braud A (2011) New roles for bacterial siderophores in metal transport and tolerance. Environ Microbiol 13:2844–2854
Shah S, Damare S (2019) Proteomic response of marine-derived: Staphylococcus cohnii #NIOSBK35 to varying Cr(VI) concentrations. Metallomics 11:1465–1471
Shah S, Damare S (2020) Cellular response of Brevibacterium casei #NIOSBA88 to arsenic and chromium—a proteomic approach. Braz J Microbiol 51:1885–1895
Shiny Matilda C, Madhusudan I, Gaurav Isola R, Shanthi C (2020) Potential of proteomics to probe microbes. J Basic Microbiol 60:471–483
Somasegaran P, Hoben HJ (1994) Handbook for Rhizobia: methods in legume-rhizobium technology. Springer-Verlag, New York, Inc., 450
Sun L, Zhu G, Dovichi NJ (2013) Comparison of the LTQ-Orbitrap Velos and the Q-Exactive for proteomic analysis of 1–1000 ng RAW 264.7 cell lysate digests. Rapid Commun Mass Spectrom 27:157–162
Tam LT, Eymann C, Albrecht D et al (2006) Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ Microbiol 8:1408–1427
Tatusov RL, Natale DA, Garkavtsev IV et al (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28
Thatoi H, Das S, Mishra J, Prasad B (2014) Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review. J Environ Manag 146:383–399
Thompson MR, VerBerkmoes NC, Chourey K et al (2007) Dosage-dependent proteome response of Shewanella oneidensis MR-1 to acute chromate challenge. J Proteome Res 6:1745–1757
Troxell B, Hassan HM (2013) Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol 3:59
Viamajala S, Peyton BM, Apel WA, Petersen JN (2002) Chromate/nitrite interactions in Shewanella oneidensis MR-1: evidence for multiple hexavalent chromium [Cr(VI)] reduction mechanisms dependent on physiological growth conditions. Biotechnol Bioeng 78:770–778
Viti C, Marchi E, Decorosi F, Giovannetti L (2014) Molecular mechanisms of Cr(VI) resistance in bacteria and fungi. FEMS Microbiol Rev 38:633–659
Völkel S, Hein S, Benker N et al (2020) How to cope with heavy metal ions: cellular and proteome-level stress response to divalent copper and nickel in Halobacterium salinarum R1 planktonic and biofilm cells. Front Microbiol 10:3056
Wagner M, Nicell JA (2002) Detoxification of phenolic solutions with horseradish peroxidase and hydrogen peroxide. Water Res 36:4041–4052
Wall T, Båth K, Britton RA et al (2007) The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl Environ Microbiol 73:3924–3935
Wang DZ, Xie ZX, Zhang SF (2014) Marine metaproteomics: current status and future directions. J Proteomics 97:27–35
Wang J-C, Ren J, Yao H-C et al (2016) Synergistic photocatalysis of Cr(VI) reduction and 4-chlorophenol degradation over hydroxylated α-Fe2O3 under visible light irradiation. J Hazard Mater 311:11–19
Wöhlbrand L, Trautwein K, Rabus R (2013) Proteomic tools for environmental microbiology—a roadmap from sample preparation to protein identification and quantification. Proteomics 13:2700–2730
Wijte D, van Baar BLM, Heck AJR, Altelaar AFM (2011) Probing the proteome response to toluene exposure in the solvent tolerant pseudomonas putida S12. J Proteome Res 10:394–403. https://doi.org/10.1021/pr100401n
Yang W, Li X, Xi D et al (2021) Synergistic chromium(VI) reduction and phenol oxidative degradation by FeS2/Fe0 and persulfate. Chemosphere 281:130957
Yung MC, Ma J, Salemi MR et al (2014) Shotgun proteomic analysis unveils survival and detoxification strategies by Caulobacter crescentus during exposure to uranium, chromium, and cadmium. J Proteome Res 13:1833–1847
Zawadzka AM, Kim Y, Maltseva N et al (2009) Characterization of a Bacillus subtilis transporter for petrobactin, an anthrax stealth siderophore. Proc Natl Acad Sci USA 106:21854–21859
Zhai Q, Xiao Y, Zhao J et al (2017) Identification of key proteins and pathways in cadmium tolerance of Lactobacillus plantarum strains by proteomic analysis. Sci Rep 7:1–17
Zhu DX, Garner AL, Galburt EA, Stallings CL (2019) CarD contributes to diverse gene expression outcomes throughout the genome of Mycobacterium tuberculosis. Proc Natl Acad Sci USA 116:13573–13581
Acknowledgements
The authors sincerely acknowledge the technical assistance of Dr. Eduardo A. Callegari and his team members. They are all from Proteomics Core Facility, in the Biomedical Research Infrastructure Network (BRIN)-Sanford School of Medicine, University of South Dakota. Besides, the authors would like to thank to Messrs. Ryan Johnson and Bill Conn from USD-IT Research Computing for their collaboration in the server maintenance and operation.
Funding
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), through Fondo para la Investigación Científica y Tecnológica (FONCyT), Argentina under Grant (PICT Start up 0018/2016). MF has a postdoctoral fellowship from CONICET. EAC and MDP integrate the SD-BRIN (NIH-NIGMS), Sanford School of Medicine, University of South Dakota. PSG and EA are researchers of CONICET (Argentine).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study investigation, design of the work and conceptualization. Material preparation and electrophoresis assays were performed by MF; proteomic analyses were carried out by EAC and MDP. Data collection and analysis were performed by MF and EAC. EA, PSG and EAC contributed with Funding acquisition and Project administration. MF wrote the original draft. All authors participated of reviewing & editing. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This research did not involve any human participants or animals. Bacterial strain used and analyzed in the present study was legally accessed, according to specifications and protocols of the Competent National Authority. Permission N°20376408874819 of the Secretary of Environment and Climate Change of Córdoba Province, Argentina.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fernandez, M., Callegari, E.A., Paez, M.D. et al. Proteomic analysis to unravel the biochemical mechanisms triggered by Bacillus toyonensis SFC 500-1E under chromium(VI) and phenol stress. Biometals 36, 1081–1108 (2023). https://doi.org/10.1007/s10534-023-00506-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-023-00506-9