Abstract
Cell migration is a fundamental biological process involved in for example embryonic development, immune system and wound healing. Cell migration is also a key step in cancer metastasis and the human copper chaperone Atox1 was recently found to facilitate this process in breast cancer cells. To explore the role of the copper chaperone in other cell migration processes, we here investigated the putative involvement of an Atox1 homolog in Caenorhabditis elegans, CUC-1, in distal tip cell migration, which is a key process during the development of the C. elegans gonad. Using knock-out worms, in which the cuc-1 gene was removed by CRISPR-Cas9 technology, we probed life span, brood size, as well as distal tip cell migration in the absence or presence of supplemented copper. Upon scoring of gonads, we found that cuc-1 knock-out, but not wild-type, worms exhibited distal tip cell migration defects in approximately 10–15% of animals and, had a significantly reduced brood size. Importantly, the distal tip cell migration defect was rescued by a wild-type cuc-1 transgene provided to cuc-1 knock-out worms. The results obtained here for C. elegans CUC-1 imply that Atox1 homologs, in addition to their well-known cytoplasmic copper transport, may contribute to developmental cell migration processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Copper (Cu) is an essential nutrient in living organisms acting as a cofactor in proteins facilitating for example respiration, iron transport, oxidative stress protection, peptide hormone production, pigmentation, and blood clotting (Puig and Thiele 2002; Matson Dzebo et al. 2016). In order to avoid toxicity of free Cu ions, intracellular Cu is regulated by devoted Cu transport proteins that assist uptake, efflux, and distribution of the metal ion to load Cu-dependent proteins (Festa and Thiele 2011; Matson Dzebo et al. 2016). In human cells, after copper transporter protein 1 (Ctr1)-mediated uptake, the cytoplasmic Cu chaperone Atox1 transports Cu to ATP7A and ATP7B in the trans-Golgi network (Puig and Thiele 2002). In support of possible redundancy, it was recently shown that glutaredoxin 1 could replace Atox1 and deliver Cu to ATP7B (Maghool et al. 2020). ATP7A/B are P1B-type ATPases that use ATP hydrolysis to transfer Cu to the lumen for loading of target Cu-dependent enzymes, such as ceruloplasmin, tyrosinase and lysyl oxidase (Matson Dzebo et al. 2016). The Atox1-ATP7A/B Cu transport pathway is conserved in many organisms, including bacteria, yeast and (of importance for this study) the round-worm Caenorhabditis elegans (C. elegans) (Klomp et al. 1997; Wakabayashi et al. 1998, Puig and Thiele 2002).
Because Cu is important as a cofactor in many proteins enabling key biological processes (Turski and Thiele 2009; Grubman and White 2014; Matson Dzebo et al. 2016), it is no surprise that Cu is required in several distinguishing cancer phenomena, and cancer patients’ serum and tumors have increased Cu levels (Denoyer et al. 2015). We recently showed that Atox1 facilitates breast cancer cell migration (Blockhuys et al. 2020) and, upon analyzing breast cancer patient tumor data, high Atox1 levels in the tumors correlated with worse prognosis of patient survival (Blockhuys et al. 2020). In addition, we found Atox1 to localize to membrane protrusions in migrating breast cancer cells (Blockhuys and Wittung-Stafshede 2017), further supporting a role for Atox1 in cell migration. Most cancer patients die from complications of metastases (i.e., spreading of the primary tumor to other organs). Since a fundamental step in metastasis is cell migration, better understanding of cell migration mechanisms and pathways are of high importance.
Cancer and organism development may share many aspects (Bernhardt et al. 2012); for example, there is also a need for Cu during pregnancy such that even temporal nutritional Cu deficiency can result in long-lasting neurological effects on the offspring (Hamza et al. 2001; Keen et al. 2003). Moreover, mice in which Atox1 was knocked out often died after birth, suggesting that Atox1 (and, thus, Cu transport) is important for appropriate embryo development (Hamza et al. 2001). When we probed Atox1 expression during early (pre-implantation) mouse embryo development, we found its expression level to increase dramatically already at the eight-cell stage and, silencing of Atox1 strongly diminished Oct4 expression in mouse embryonic cells (Celauro et al. 2017). The Oct4 protein is a master transcription factor that regulates pluripotency and differentiation; notably, it controls an extensive gene network that drives the maternal-to-embryo transition (Zuccotti et al. 2011).
To probe the hypothesis that Atox1, and its homologs, may play yet unexplored roles in developmental cell migration processes, we here turned to the model organism C. elegans. This is an excellent model system where at least 40% of human genes are conserved (Jorgensen and Mango 2002; Rual et al. 2004); moreover, the animals are easy to grow, have short life span and are transparent. The latter property allows for observation of individual cells and subcellular details during animal development using differential interference contrast (DIC) microscopy. Homologs to Atox1 and ATP7A/B in worms have been identified, named CUC-1 and CUA-1, respectively, and have demonstrated Cu transport functions (Wakabayashi et al. 1998; Chun et al. 2017). Notably, CUA-1 exhibits Cu-dependent redistribution in cells, similar to ATP7A/B in humans (Chun et al. 2017). CUC-1 has 39.1% sequence identity to Atox1 (Fig. 1) and could replace yeast Atx1 as a Cu chaperone in Atx1-deficient yeasts (Wakabayashi et al. 1998). However, in contrast to humans, where Atox1 is expressed ubiquitously, C. elegans CUC-1 showed tissue-specific expression, specifically found in intestinal cells of adult worms and hypodermal cells in the larvae (Wakabayashi et al. 1998; Chun et al. 2017).
Amino acid sequence alignment of Atox1 homologs. The full amino-acid sequence of CUC-1 was aligned with full sequences of yeast Atx1p and human Atox1. Boxed residues are conserved in all three proteins. Residues in red are identical to CUC-1 in one or both other proteins. The top two boxes include the conserved MTCXGC Cu-binding motif (with the two cysteines coordinating the Cu ion)
During post-embryonic C. elegans development, the morphology of the hermaphroditic gonad is determined. The somatic gonad consists of different tissues, each with specific functions and distinct anatomical features, that are intimately associated with the germ line and have critical roles in development, organization, and function in adult worms. The process determining the gonad takes place during larvae stages L2 to L4 and involves appropriate migration of two specialized leader cells, the so-called distal tip cells (DTCs). At the larval stage L2, the DTCs migrate away from the gonad primordium. At L3, the DTCs turn and migrate to the dorsal side, followed by a U-turn and migration towards the mid-body in the L4 stage (Fig. 2a). Migration ceases opposite to the vulva, and with the two U-shaped gonad arms in place; worms are defined as adults when the vulva completes its development. DTCs are particularly amenable to in situ cell migration analysis because (in contrast to most vertebrate tissue) C. elegans is transparent. Moreover, the C. elegans genome is conserved, which allows translation of functions to other systems, including humans. Dysfunctional DTC migration may result in a range of gonad development defects, such as turning, pathfinding and cessation problems which are easily detected under a microscope (Fig. 2b). Many genes have been identified as crucial for the DTC migration process in worms, including for example Rho GTPases, integrins and metalloproteases (Lundquist et al. 2001; Lee et al. 2005; Wong and Schwarzbauer 2012), but no Cu dependence has been noted.
a Scheme of the U-shaped migratory path of the distal tip cells (green) that shape the hermaphrodite gonad arms during worm larval development; Phase 1: larval stage 2 (red), phase 2: larval stage 3 (yellow) and phase 3: larval stage 4 (blue). b Patterns of DTC migration defects. Arrow shows migration direction. a normal U-shaped migratory path, b and c turning defects (b, ventralized migration; c, wrong turn), d pathfinding defects (change in direction), e cessation problems (overshoot of migration)
In this study, taking advantage of the DTC migration in situ model, we have characterized C. elegans with and without the cuc-1 gene to reveal a putative role of CUC-1 in the DTC migration process. We found that upon knock-out of the cuc-1 gene, C. elegans animals exhibit defective DTC migration patterns (along with decreased brood size) that can be rescued by wild-type cuc-1 provided as a transgene. Our results suggest that CUC-1 is a co-regulator of DTC migration during C. elegans larvae development.
Materials and methods
C. elegans worm cultures and strains
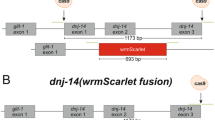
Caenorhabditis elegans was cultivated at 20 °C on nematode growth medium (NGM) plates with E. coli OP50 (worm base ID WBStrain00041969) as the food source. E. coli OP50 is an uracil auxotroph whose growth is limited on NGM plates. A limited bacterial lawn is desirable because it allows for easier observation and better mating of the worms. The media, culture and handling of C. elegans follow what was described by Brenner (1974). We used two different C. elegans strains. The wild-type C. elegans strain N2 was provided by the Caenorhabditis Genetics Center (funded by the NIH National Center for Research Resources, NCRR), and the PHX1006(cuc-1(syb1006)/hT2)) strain was created by Suny Biotech (Fuzhou City, China) using CRISPR/Cas9 to delete cuc-1 (deletion site shown in Fig. 3a). The deletion of the cuc-1 gene was confirmed by PCR using the following primers: 5′-AATGGCTTTCGGACTACTGT-3′ and 5′- CTCACCACATCGTTAGTAGG-3′. For Cu complementation, CuCl2 was added at a concentration of 50 µM when preparing NGM agar dishes. NGM mixture itself does not contain Cu, however, the E. coli OP50 and the yeast extract in the agar may supply Cu to the worms.
Growth assay
Synchronized worms at L1 stage from both N2 worms (wild type) and cuc-1 mutant stocks were plated onto NGM plates seeded with OP50. For the growth rate assay, 20–25 worms were mounted and photographed by DIC microscope when worms were grown for 24, 48, 72 and 96 h. Worm length (excluding the thin tail tip) was measured using the ImageJ software (Schindelin et al. 2012).
Brood size assay
Synchronous L1 worms were plated onto NGM plates with OP50 and after an additional 48 h of growth (to reach L4 stage), 10 worms were singled out onto new plates. The worms were transferred daily during the fertile period and live progeny were counted 3 days after removal of the hermaphrodite.
Life span assay
Synchronous L4 worms were plated in groups of five onto NGM plates with OP50. 100 worms were picked for both N2 and cuc-1 mutant strains. The worms were transferred every second day during the fertile period and once a week thereafter. All worms were monitored every day and scored as dead when failing to respond upon several touches on the head with the worm-pick.
Scoring DTC migration defect phenotypes
Synchronous L1 worms were plated onto NGM plates with OP50, and after an additional 72 h when grown to young adults, hermaphrodites were observed on 2% agar pads using a Zeiss Axio photo microscope equipped with a 60X objective and Nomarski optics (Sulston and Horvitz 1977). 40 worms were analyzed in each experiment, and the experiment was repeated three times. The trajectories of the DTCs were manually deduced from inspections of the gonad arms. Animals were observed with anterior arms on the left and posterior arms on the right side. Both sides were scored.
Construction of wild-type cuc-1 rescue plasmid
The pCUC-1 rescue construct was generated in-house with a Gibson assembly kit (Gibson Assembly® Cloning Kit (New England Biolabs)) using the following primers.
-
1.
5′- GGAATTCGCCCTTGTCTAGAGTAAGCTATTTTAGAGATTTTTGAC-3′ and 5′- TATCTGCAGAATTCGCCCTTACGTTTACCTTTCTGAGAAG − 3′ to amplify the cuc-1 gene including 2 kb of upstream regulatory sequence using N2 worm genomic DNA as template.
-
2.
5′-AAGGGCGAATTCTGCAGATATCCATCACACTGGCGGCCGCTCGA-3′ and 5′- TCTAGACAAGGGCGAATTCCAGCACACTGGCGGCCGTTACTAGTT-3′ as vector primers amplified from pPAQR-2::GFP construct (Svensson et al. 2011).
The plasmid construct was confirmed by sequencing using 5′-GTGGCGGCCGCTCTAGAACTAGTAACGGCCGCCAGT-3′ and 5′-TAGATGCATGCTCGAGCGGCCGCCAGTGTGATGGA-3′. The assembled pCUC-1 plasmid was injected into PHX1006 (cuc-1(syb1006)/hT2) worms at 10 ng/µl together with 5 ng/µl pPD118.33 (Pmyo-2::GFP) (Addgene plasmid #1596) and 85 ng/µl pBSKS (Stratagene).
Statistics
The Student’s t-tests were used to determine statistical significance of our measures. All experiments were repeated at least three times. Asterisks presented in the figures indicate various degrees of significance, as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
Results
C. elegans worms with the cuc-1 gene deleted (cuc-1 mutant) using CRISPR-Cas9 technology (Fig. 3a) were tested with PCR (Fig. 3b) to assure successful gene deletion. Importantly, the deletion was not lethal to worms. To determine the effects of the cuc-1 mutation on worm properties, C. elegans worms were grown on NGM plates fed with the E. coli strain OP50 over 20 days. The worm growth assay showed that cuc-1 mutant worms exhibited wild-type length during development except for the L4 to young adult stage, where a ~ 10% reduction in length was noted (Fig. 4a). Importantly, the cuc-1 mutant displayed a 50% reduction in brood size compared to wild-type worms (Fig. 4b), and the cuc-1 mutant also showed a somewhat shorter life span (p = 0.00007) than wild-type worms (Fig. 4c).
Characterization of cuc-1 mutant versus wild-type (N2) C. elegans at 24, 48, 72 and 96 h of incubation post L1 stage (condition with no supplemental Cu). a Growth assay (ngroup=25). b Brood size assay (ngroup=10). c Life span assay (ngroup=100). d Examples of DTC migration defects found in cuc-1 mutant C. elegans. Asterisks indicate vulva. White dashed lines indicate gonad shapes. The scale bar in wild-type image represents 50 µm for all images. e Quantification of DTC migration defects in cuc-1 mutant C. elegans (ngroup=40; three independent groups analyzed). Statistical significance determined using the Student´s t-test with * for p < 0.05, ** for p < 0.01, *** for p < 0.001
Next, we analyzed the DTC migration patterns by scoring wild-type and cuc-1 mutant worms at the young adult stage. Upon analysis of defects, we found that almost 10% of cuc-1 mutants had DTC migration defects (Fig. 4 d,e), whereas no such defects were observed in wild-type worms. Table 1 reports the types of DTCs migration defects (Fig. 2b) observed, revealing that most defects (in more than half of defective cuc-1 mutant worms) were classified as pathfinding defects.
The normal growth condition for the worms (NGM petri plates with E. coli OP50 food) does not include added Cu, although the E. coli and the yeast extract in the agar are sources of Cu and thus the worms have access to some Cu (but exact concentrations are not known). Previous studies have shown that removal of all Cu by addition of chelation compounds, or addition of excessive levels of Cu to the growth media, diminish growth of C. elegans, but for up to 100 µM of Cu supplemented in the NGM agar, there are no growth defects noted in worms (Chun et al. 2017). Therefore, to test whether cuc-1 mutants are sensitive to the level of Cu, we compared the phenotypes of wild-type and cuc-1 mutants grown on normal plates or NGM plates supplemented with 50 µM Cu. Thus, although both conditions will contain Cu, the latter should give the worms access to more Cu.
At the 50 µM Cu supplemented condition, we found again a 50% reduction of brood size and shorter worm lengths at the L4 to adult stages for the cuc-1 mutant worms; in addition, for this condition we also found shorter worm lengths at L3 to L4 stages for the cuc-1 mutant worms (Fig. 5a, b). The life span assay showed again a somewhat reduced life span for cuc-1 mutant worms as compared to wild-type worms (Fig. 5c). In the Cu-supplemented condition, 15% of cuc-1 mutant worms exhibited DTC migration defects (Fig. 5d, e) and again pathfinding defects dominated (Table 1). Importantly, DTCs migration in wild-type worms remained unaffected at the Cu-supplemented condition. The similar outcomes for the two different Cu conditions imply that the observed defects due to cuc-1 deletion is not sensitive to variation in external Cu supply. We note that we have not assessed Cu content inside the worms (e.g., in the DTCs) which, due to regulation of cellular Cu uptake and transport in worms (Wakabayashi et al. 1998; Chun et al. 2017), may be rather similar at the two conditions.
Characterization of cuc-1 mutant versus wild-type (N2) C. elegans at 24, 48, 72 h incubation post L1 stage (condition with 50 µM supplemental Cu). a Growth assay (ngroup=25). b Brood size assay (ngroup=10). c Life span assay (ngroup=100). d Examples of DTC migration defects found in cuc-1 mutant C. elegans. Asterisks indicate vulva. White dashed lines indicate gonad shapes. The scale bar in wild-type image represents 50 µm for all images. e Quantification of DTC migration defects for cuc-1 mutant C. elegans. (ngroup=40; three independent groups analyzed). Statistical significance determined using the Student´s t-test with * for p < 0.05, ** for p < 0.01, *** for p < 0.001
To prove that the observed DTC migration defects are due to absence of the CUC-1 protein, and not due to indirect effects of the gene deletion, we injected a plasmid containing the wild-type cuc-1 gene (named pCUC-1) into cuc-1 mutant worms. First, pCUC-1 was injected into the gonad syncytium of cuc-1 mutant worms to establish a transgenic line. Transgenic worms in subsequent generations were verified by PCR (Fig. 6a). Scoring of ‘rescued’ worms at the young adult stage showed no DTC migration defects (Fig. 6b), whereas, as before, around 10% of the non-transgenic cuc-1 mutant worms investigated in parallel exhibited DTC migration defects (Fig. 6c). Thus, by adding back the cuc-1 gene, cuc-1 mutant worms are rescued with respect to migration of the DTCs. This demonstrates that the migration defects observed in cuc-1 mutants are due to lack of CUC-1 protein.
a PCR data show the cuc-1 gene to be successfully inserted into cuc-1 mutant worms upon transgene plasmid microinjection. b Examples of DTC migration in rescued cuc-1 mutant (C) elegans (ngroup=40; three independent groups analyzed). Asterisks indicate vulva. White dashed lines indicate gonad shapes. No DTC migration defects were detected. The scale bar represents 50 µm. c Quantification of DTC migration defects for rescued cuc-1 mutant and non-rescued mutant C. elegans worms analyzed in parallel. Statistical significance determined using the Student´s t-test with * for p < 0.05, ** for p < 0.01, *** for p < 0.001
Discussion
Gonad morphogenesis in C. elegans is dependent on specific migration patterns of DTCs and, for many reasons, is an excellent system to dissect the molecular basis of developmental cell migration (Lee et al. 2005). Several proteins and pathways have been related to the distinct steps of DTC migration that eventually results in two U-shaped gonad arms (Lundquist et al. 2001; Lee et al. 2005). Because the human Cu chaperone Atox1 has been found to facilitate cancer cell migration (Blockhuys et al. 2020) and play an apparent role in pre-implantation mouse embryo development (Celauro et al. 2017), we here set out to test the role of the C. elegans Atox1 homolog, CUC-1, in DTC migration.
It has been reported that CUC-1, together with CUA-1 (which is the C. elegans homolog to human ATP7A/B), function in Cu transport as in humans, but with tissue-specific expression (Wakabayashi et al. 1998; Chun et al. 2017). Here, we investigated C. elegans with the cuc-1 gene removed using CRISPR-Cas9 technology. In similarity to a study in which the cuc-1 gene was silenced using RNAi (Chun et al. 2017), we found the mutated worms to display reduced brood size. Because the deletion of the gene did not affect worm growth and life span much, we were able to probe migration of DTCs during larval development as a function of CUC-1 presence.
We found that whereas wild-type worms had no DTC migration defects at the two conditions studied, approximately 10% (normal growth condition) and 15% (50 µM Cu supplemented to the plates) of cuc-1 mutant worms showed DTC migration defects. When the defects were classified (at both conditions), we found pathfinding problems to be most common followed by turning problems. However, our classification gives only rough estimates as, in some cases, more than one problem may be present, and the total number of defective worms analyzed are small. The results for CUC-1 deletion should be placed in relation to similar studies investigating the roles of other gene products in DTC migration. Reporting migration defects as a percentage of worms with defects is a common way to quantify dysfunction due to gene mutations. In other studies, the percentage of worms with DTC migration defects range from around 10–20% defective worms to much higher percentages, depending on the gene deleted. Rho GTPases such as CED-10 and MIG-2 are known to play key roles in DTC migration (Lundquist et al. 2001; Lee et al. 2005; Wong and Schwarzbauer 2012) and are regulated by many additional proteins (Demarco and Lundquist 2010; Tannoury et al. 2010). In similarity to the detected magnitudes of defects here for cuc-1 deletion, mutation in the mig-2 and ced-10 genes resulted in DTC migration defects (with pathfinding problems highlighted) in 27% and 12% of worms, respectively (Lundquist et al. 2001). Nonetheless, due to the low percentage of worms affected by cuc-1 deletion, it may be speculated that there is functional redundancy such that other proteins can substitute (in part) for CUC-1.
We note that the observation for CUC-1 is not a general result of removing a Cu-binding protein from the organism, as it was reported that gene knockdown of the Cu-binding protein CUTC-1 (an ortholog of human CUTC) in C. elegans did not affect brood size, worm length, or DTC migration at added Cu concentrations up to 100 µM (Calafato et al. 2008). Instead, CUTC-1 was concluded to protect worms from toxic effects of excess Cu (above 100 µM).
Our findings demonstrate that the Atox1 homolog in worms, CUC-1, plays a role in promoting correct DTC migration during C. elegans development. Thus, CUC-1 should be considered a new modulator of the extensive network of proteins that facilitate DTCs migration in worms. CUC-1 may play Cu-dependent or Cu-independent roles in DTC migration. We speculate that there may be unknown Cu dependencies for other proteins in these processes to which CUC-1 would deliver Cu. Speculatively, Cu-loaded CUC-1 may help mediate oxidation/reduction processes that regulate Rho GTPases implicated in DTC migration, such as Rac, Rho, and Cdc-42. In human cells, Atox1 was found to mediate cancer cell migration by (via Cu transport) promoting extracellular, Cu-dependent, collagen-remodeling lysyl oxidase (LOX) activity. C. elegans worms do not have LOX proteins but instead depend on other proteins that regulate the extracellular matrix. No Cu dependence for those proteins have been reported but, nonetheless, many are oxidases, peroxidases and thioredoxin-like proteins (Myllyharju and Kivirikko 2004) that may turn out to depend on Cu. There are also possible Cu-independent mechanisms responsible for the observations: CUC-1 may transmit signals via protein-protein interactions that may be dependent on redox status of CUC-1’s two cysteine residues (in the Cu-binding loop). Such redox properties have been reported for Atox1 (Hatori and Lutsenko 2013) and may be conserved in CUC-1. Taken together, Atox1 and its homologs (e.g., CUC-1) appear to have functions, beyond that of pure cytoplasmic Cu transport, that aid in cell migration during organism development.
Data availability
All data and material are available upon request from the corresponding author.
References
Bernhardt M, Galach M, Novak D, Utikal J (2012) Mediators of induced pluripotency and their role in cancer cells—current scientific knowledge and future perspectives. Biotechnol J 7(6):810–821
Blockhuys S, Brady DC, Wittung-Stafshede P (2020) Evaluation of copper chaperone ATOX1 as prognostic biomarker in breast cancer. Breast Cancer 27(3):505–509
Blockhuys S, Wittung-Stafshede P (2017) Copper chaperone Atox1 plays role in breast cancer cell migration. Biochem Biophys Res Commun 483(1):301–304
Blockhuys S, Zhang X, Wittung-Stafshede P (2020) Single-cell tracking demonstrates copper chaperone Atox1 to be required for breast cancer cell migration. Proc Natl Acad Sci USA 117(4):2014–2019
Brenner S (1974) The genetics of Caenorhabditis elegans . Genetics 77(1):71–94
Calafato S, Swain S, Hughes S, Kille P, Sturzenbaum SR (2008) Knock down of Caenorhabditis elegans cutc-1 exacerbates the sensitivity toward high levels of copper. Toxicol Sci 106(2):384–391
Celauro E, Mukaj A, Fierro-Gonzalez JC, Wittung-Stafshede P (2017) Copper chaperone ATOX1 regulates pluripotency factor OCT4 in preimplantation mouse embryos. Biochem Biophys Res Commun 491(1):147–153
Chun H, Sharma AK, Lee J, Chan J, Jia S, Kim BE (2017) The intestinal copper exporter CUA-1 Is required for systemic copper homeostasis in Caenorhabditis elegans. J Biol Chem 292(1):1–14
Demarco RS, Lundquist EA (2010) RACK-1 acts with Rac GTPase signaling and UNC-115/abLIM in Caenorhabditis elegans axon pathfinding and cell migration. PLoS Genet 6(11):e1001215
Denoyer D, Masaldan S, La Fontaine S, Cater MA (2015) Targeting copper in cancer therapy: ‘Copper That Cancer’’.’ Metallomics 7(11):1459–1476
Festa RA, Thiele DJ (2011) Copper: an essential metal in biology. Curr Biol 21(21):R877–R883
Grubman A, White AR (2014) Copper as a key regulator of cell signalling pathways. Expert Rev Mol Med 16:e11
Hamza I, Faisst A, Prohaska J, Chen J, Gruss P, Gitlin JD (2001) The metallochaperone Atox1 plays a critical role in perinatal copper homeostasis. Proc Natl Acad Sci USA 98(12):6848–6852
Hatori Y, Lutsenko S (2013) An expanding range of functions for the copper chaperone/antioxidant protein Atox1. Antioxid redox Signal 19(9):945–957
Jorgensen EM, Mango SE (2002) The art and design of genetic screens: Caenorhabditis elegans. Nat Rev Genet 3(5):356–369
Keen CL, Hanna LA, Lanoue L, Uriu-Adams JY, Rucker RB, Clegg MS (2003) Developmental consequences of trace mineral deficiencies in rodents: acute and long-term effects. J Nutr 133(5 Suppl 1):1477S-1480S
Klomp LW, Lin SJ, Yuan DS, Klausner RD, Culotta VC, Gitlin JD (1997) Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J Biol Chem 272(14):9221–9226
Lee M, Shen B, Schwarzbauer JE, Ahn J, Kwon J (2005) Connections between integrins and Rac GTPase pathways control gonad formation and function in C. elegans. Biochim Biophys Acta 1723(1–3):248–255
Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI (2001) Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance cell migration apoptotic cell phagocytosis. Development 128(22):4475–4488
Maghool S, Fontaine SL, Roberts BR, Kwan AH, Maher MJ (2020) Human glutaredoxin-1 can transfer copper to isolated metal binding domains of the P1B-type ATPase, ATP7B. Sci Rep 10(1):4157
Matson Dzebo M, Arioz C, Wittung-Stafshede P (2016) Extended functional repertoire for human copper chaperones. Biomol Concepts 7(1):29–39
Myllyharju J, Kivirikko KI (2004) Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet 20(1):33–43
Puig S, Thiele DJ (2002) Molecular mechanisms of copper uptake and distribution. Curr Opin Chem Biol 6(2):171–180
Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M (2004) Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14(10B):2162–2168
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682
Sulston JE, Horvitz HR (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56(1):110–156
Svensson E, Olsen L, Morck C, Brackmann C, Enejder A, Faergeman NJ, Pilon M (2011) The adiponectin receptor homologs in C. elegans promote energy utilization and homeostasis. PLoS ONE 6(6):e21343
Tannoury H, Rodriguez V, Kovacevic I, Ibourk M, Lee M, Cram EJ (2010) CACN-1/Cactin interacts genetically with MIG-2 GTPase signaling to control distal tip cell migration in C. elegans. Dev Biol 341(1):176–185
Turski ML, Thiele DJ (2009) New roles for copper metabolism in cell proliferation, signaling, and disease. J Biol Chem 284(2):717–721
Wakabayashi T, Nakamura N, Sambongi Y, Wada Y, Oka T, Futai M (1998) Identification of the copper chaperone, CUC-1, in Caenorhabditis elegans: tissue specific co-expression with the copper transporting ATPase, CUA-1. FEBS Lett 440(1–2):141–146
Wong MC, Schwarzbauer JE (2012) Gonad morphogenesis and distal tip cell migration in the Caenorhabditis elegans hermaphrodite. Wiley Interdiscip Rev Dev Biol 1(4):519–531
Zuccotti M, Merico V, Bellone M, Mulas F, Sacchi L, Rebuzzini P, Prigione A, Redi CA, Bellazzi R, Adjaye J, Garagna S (2011) Gatekeeper of pluripotency: a common Oct4 transcriptional network operates in mouse eggs and embryonic stem cells. BMC Genome 12:1–13
Acknowledgements
Open access funding provided by Chalmers University of Technology.
Funding
We thank the Knut and Alice Wallenberg foundation, the Swedish Research Council and the Swedish Cancer Foundation for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflcit of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, X., Blockhuys, S., Devkota, R. et al. The Caenorhabditis elegans homolog of human copper chaperone Atox1, CUC-1, aids in distal tip cell migration. Biometals 33, 147–157 (2020). https://doi.org/10.1007/s10534-020-00239-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-020-00239-z