Abstract
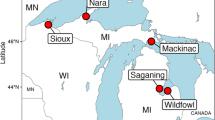
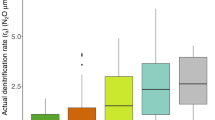
Large lakes can efficiently remove reactive nitrogen (N) through denitrification, but nitrate levels in some large oligotrophic lakes are increasing, indicating that denitrification in these lakes is not capable of removing excess N. To better understand how lake trophic status and sediment redox conditions affect the capacity of the microbial community to remove excess N, we measured potential denitrification rates at 86 different stations across Lakes Superior, Huron, Erie, and Ontario. We also relate sediment microbial communities to potential denitrification rates and sediment characteristics for a subset of these sites. In eutrophic/mesotrophic Lake Erie, characterized by sediment with minimal oxygen penetration and relatively high sediment carbon (C) and N, potential denitrification rates were relatively high and increased by 2–3 orders of magnitude in response to additional nitrate and organic C. In contrast, in oligotrophic Lakes Superior and Ontario, and mesotrophic Lake Huron, where oxygen can penetrate several cm into sediment, potential denitrification rates were generally low and did not respond to additional nitrate and organic carbon. Sediment microbial communities showed a similar pattern across this gradient, correlated with potential denitrification rates, sediment %C, and bottom-water nitrate concentrations. This observed relationships between sediment redox conditions, potential denitrification rates, and microbial diversity suggest that sediment microbial communities in these and other oligotrophic large lakes may already be operating at or near their maximum denitrification rates. Unlike mesotrophic Lake Erie, microbial communities in oligotrophic lake sediments may lack the ability to mitigate increases in N loading through denitrification.









Similar content being viewed by others
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Bennington V, McKinley GA, Urban NR, McDonald CP (2012) Can spatial heterogeneity explain the perceived imbalance in Lake Superior’s carbon budget? A model study. J Geophys Res 117:G03020
Bollmann A, Bullerjahn GS, McKay RM (2014) Abundance and diversity of ammonia-oxidizing archaea and bacteria in sediments of trophic end members of the Laurentian Great Lakes, Erie and Superior. PLoS One 9:e97068
Bruesewitz DA, Tank JL, Hamilton SK (2012) Incorporating spatial variation of nitrification and denitrification rates into whole-lake nitrogen dynamics. J Geophys Res 117:G00N07
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach, 2nd edn. Springer-Verlag, New York
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JG, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Purnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010a) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen TGL, Knight R (2010b) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108:4516–4522
Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M (2015) Complete nitrification by Nitrospira bacteria. Nature. doi:10.1038/nature16461
Davidson EA, Seitzinger S (2006) The enigma of progress in denitrification research. Ecol Appl 16:2057–2063
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 72:5069–5072
Dodla SK, Wang JJ, DeLaune RD, Cook RL (2008) Denitrification potential and its relation to organic carbon quality in three coastal wetland soils. Sci Total Environ 407:471–480
Dolan DM, Chapra SC (2011) Great lakes total phosphorus loads and models: a fifteen year update. U.S. EPA, Final Report, Grant no. GL 00E58501
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE (2007) Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett 10:1135–1142
Eyre BD, Maher DT, Squire P (2013) Quantity and quality of organic matter (detritus) drives N2 effluxes (net denitrification) across seasons, benthic habitats, and estuaries. Glob Biogeochem Cycles 27:1083–1095
Finlay JC, Small GE, Sterner RW (2013) Human influences on nitrogen removal in lakes. Science 342:247–250
Groffman PM, Altabet MA, Böhlke JK, Butterbach-Bahl K, David MB, Firestone MK, Giblin AE, Kana TM, Nielsen LP, Voytek MA (2006) Methods for measuring denitrification: diverse approaches to a difficult problem. Ecol Appl 16:2091–2122
Harrison JA, Maranger RJ, Alexander RB, Giblin AE, Jacinthe PA, Mayorga E, Seitzinger SP, Sobota DJ, Wollheim WM (2009) The regional and global significance of nitrogen removal in lakes and reservoirs. Biogeochemistry 93:143–157
Heylen K, Gevers D, Vanparys B, Wittebolle L, Geets J, Boon N, De Vos P (2006) The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ Microbiol 8:2012–2021
Howarth RW, Marino R (2006) Nitrogen as the limiting nutrient for eutrophication in coastal marine ecosystems: evolving views over three decades. Limnol Oceanogr 51:364–376
Knuth ML, Kelly JR (2011) Denitrification rates in a Lake Superior coastal wetland. Aquat Ecosyst Health 14:414–421
Li J, Katsev S (2014) Nitrogen cycling in deeply oxygenated sediments: results in Lake Superior and implications for marine sediments. Limnol Oceanogr 59:465–481
Li J, Crowe SA, Miklesh D, Kistner M, Canfield DE, Katsev S (2012) Carbon mineralization and oxygen dynamics in sediments with deep oxygen penetration, Lake Superior. Limnol Oceanogr 57:1634–1650
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microb 73:1576–1585
McCrackin ML, Elser JJ (2010) Atmospheric nitrogen deposition influences denitrification and nitrous oxide production in lakes. Ecology 91:528–539
McCrackin ML, Elser JJ (2012) Denitrification kinetics and denitrifier abundances in sediments of lakes receiving atmospheric nitrogen deposition (Colorado, USA). Biogeochemistry 108:39–54
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618
McMurdie PJ, Holmes S (2013) phyloseq: an R Package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4):e61217
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10(4):e1003531
Mulholland PJ, Helton AM, Poole GC, Hall RO, Hamilton SK, Peterson BJ, Tank JL, Ashkenas LR, Cooper LW, Dahm CN, Dodds WK, Findlay SEG, Gregory SV, Grimm NB, Johnson SL, McDowell WH, Meyer JL, Vallett HM, Webster JR, Arango CP, Beaulieu JJ, Bernot MJ, Burgin AJ, Crenshaw CL, Johnson LT, Niederlehner BR, O’Brien JM, Potter JD, Sheibley RW, Sobota DJ, Thomas SM (2008) Stream denitrification across biomes and its response to anthropogenic nitrate loading. Nature 452:202–205
Price M, Dehal P, Arkin A (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490
R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Revsbech NP (1989) An oxygen microsensor with a guard cathode. Limnol Oceanogr 34:474–478
Richardson WB, Strauss EA, Bartsch LA, Monroe EM, Cavanaugh JC, Vingum L, Soballe DM (2004) Denitrification in the Upper Mississippi River: rates, controls, and contribution to nitrate flux. Can J Fish Aquat Sci 61:1102–1112
Robertson DM, Saad DA (2011) Nutrient inputs to the Laurentian Great Lakes by source and watershed estimated using Sparrow watershed models. J Am Water Resour Assoc 47:1011–1033
Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-García C, Rodríguez G, Massol-Deyá A, Krishnani KK, Ritalahti KM, Nissen S, Konstantinidis KT, Löffler FE (2012) Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci USA 109:19709–19714
Schlesinger WH (2009) On the fate of anthropogenic nitrogen. Proc Natl Acad Sci USA 106:203–208
Scott JT, McCarthy MJ (2010) Nitrogen fixation may not balance the nitrogen pool in lakes over timescales relevant to eutrophication management. Limnol Oceanogr 55:1265–1270
Seitzinger SP (1988) Denitrification in freshwater and coastal marine ecosystems: ecological and geochemical significance. Limnol Oceanogr 33:702–724
Seitzinger S, Harrison JA, Böhlke JK, Bouwman AF, Lowrance R, Peterson B, Tobias C, van Drecht G (2006) Denitrification across landscapes and waterscapes: a synthesis. Ecol Appl 16:2064–2090
Small G, Cotner JB, Finlay JC, Stark RA, Sterner RW (2013) Nitrogen transformations at the sediment-water interface across redox gradients in the Laurentian Great Lakes. Hydrobiologia 731:95–108
Small GE, Sterner RW, Finlay JC (2014) An ecological network analysis of nitrogen cycling in the Laurentian Great Lakes. Ecol Model 293:150–160
Stein LY, Klotz MG (2011) Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem Soc Trans 39:1826–1831
Stelzer RS, Scott JT, Bartsch LA, Parr TB (2014) Particulate organic matter quality influences nitrate retention and denitrification in stream sediments: evidence from a carbon burial experiment. Biogeochemistry 119:387–402
Sterner RW, Anagnostou E, Brovold S, Bullerjahn GS, Finlay JC, Kumar S, McKay RML, Sherrell RM (2007) Increasing stoichiometric imbalance in North America’s largest lake. Geophys Res Lett 34:L10406. doi:10.1029/2006GL028861
van Kessel JF (1977) Factors affecting the denitrification rate in two water sediment systems. Water Res 11:259–267
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 73:5261–5267
Wei W, Isobe K, Nishizawa T, Zhu L, Shiratori Y, Ohte N, Koba K, Otsuka S, Senoo K (2015) Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. doi:10.1038/ismej.2015.9
Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, Angenent LT, Knight R, Ley RE (2012) Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J 6:94–103
Wollheim WM, Vorosmarty CJ, Bouwman AF, Green P, Harrison J, Linder E, Peterson BJ, Seitzinger SP, Syvitski JPM (2008) Global N removal by freshwater aquatic systems using a spatially distributed, within-basin approach. Glob Biogeochem Cycles 22:2026
Yoshinara T, Knowles R (1976) Acetylene inhibition of nitrous oxide reductase by denitrifying bacteria. Biochem Biophys Res Commun 69:705–710
Acknowledgments
We thank the crew of the R/V Blue Heron for their support during this study, and the U.S. Coast Guard, Canadian Coast Guard, and Environment Canada for providing additional sampling opportunities. This material is based upon work supported by the National Science Foundation under Grants OCE-0927512 and OCE-0927277. The work conducted by the U.S. Department of Energy Joint Genome Institute was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231 and Community Sequencing Project 723.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jennifer Leah Tank.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Small, G.E., Finlay, J.C., McKay, R.M.L. et al. Large differences in potential denitrification and sediment microbial communities across the Laurentian great lakes. Biogeochemistry 128, 353–368 (2016). https://doi.org/10.1007/s10533-016-0212-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10533-016-0212-x