Abstract
Pseudomonas putida CSV86 utilizes naphthalene (Nap), salicylate (Sal), benzyl alcohol (Balc), and methylnaphthalene (MN) preferentially over glucose. Methylnaphthalene is metabolized by ring-hydroxylation as well as side-chain hydroxylation pathway. Although the degradation property was found to be stable, the frequency of obtaining Nap−Sal−MN−Balc− phenotype increased to 11% in the presence of curing agents. This property was transferred by conjugation to Stenotrophomonas maltophilia CSV89 with a frequency of 7 × 10−8 per donor cells. Transconjugants were Nap+Sal+MN+Balc+ and metabolized MN by ring- as well as side-chain hydroxylation pathway. Transconjugants also showed the preferential utilization of aromatic compounds over glucose indicating transfer of the preferential degradation property. The transferred properties were lost completely when transconjugants were grown on glucose or 2YT. Attempts to detect and isolate plasmid DNA from CSV86 and transconjugants were unsuccessful. Transfer of degradation genes and its subsequent loss from the transconjugants was confirmed by PCR using primers specific for 1,2-dihydroxynaphthalene dioxygenase and catechol 2,3-dioxygenase (C23O) as well as by DNA–DNA hybridizations using total DNA as template and C23O PCR fragment as a probe. These results indicate the involvement of a probable conjugative element in the: (i) metabolism of aromatic compounds, (ii) ring- and side-chain hydroxylation pathways for MN, and (iii) preferential utilization of aromatics over glucose.




Similar content being viewed by others
References
Anderson DG, McKay LL (1983) Simple and rapid method for isolating large plasmid DNA from lactic Streptococci. Appl Environ Microbiol 46:549–552
Basu A, Phale PS (2006) Inducible uptake and metabolism of glucose by the phosphorylative pathway in Pseudomonas putida CSV86. FEMS Microbiol Lett 259:311–316
Basu A, Apte SK, Phale PS (2006) Preferential utilization of aromatic compounds over glucose by Pseudomonas putida CSV86. Appl Environ Microbiol 72:2226–2230
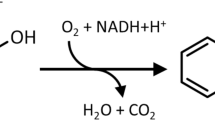
Basu A, Dixit SS, Phale PS (2003) Metabolism of benzyl alcohol via catechol ortho-pathway in methylnaphthalene-degrading Pseudomonas putida CSV86. Appl Microbiol Biotechnol 62:579–585
Burrus V, Pavlovic G, Decaris B, Guedon G (2002) Conjugative transposons: the tip of the iceberg. Mol Microbiol 46:601–610
Chakrabarty AM (1972) Genetic basis of the biodegradation of salicylate in Pseudomonas. J Bacteriol 112:815–823
Curiale MS, Mills D (1982) Integration and partial excision of a cryptic plasmid in Pseudomonas syringae pv. phaseolicola. J Bacteriol 152:797–802
Davies JI, Evans WC (1964) Oxidative metabolism of naphthalene by soil pseudomonads. The ring-fission mechanism. Biochem J 91:251–261
Dennis JJ (2005) The evolution of IncP catabolic plasmids. Curr Opin Biotechnol 16:291–298
Dennis JJ, Zylstra GJ (2004) Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J Mol Biol 341:753–768
Dunn NW, Gunsalus IC (1973) Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol 114:974–979
El-Mansi M, Anderson KJ, Inche CA, Knowles LK, Platt DJ (2000) Isolation and curing of the Klebsiella pneumoniae large indigenous plasmid using sodium dodecyl sulphate. Res Microbiol 151:201–208
Friello DA, Mylroie JR, Gibson DT, Rogers JE, Chakrabarty AM (1976) XYL, a nonconjugative xylene-degradative plasmid in Pseudomonas Pxy. J Bacteriol 127:1217–1224
Fujii T, Takeo M, Maeda Y (1997) Plasmid-encoded genes specifying aniline oxidation from Acinetobacter sp. strain YAA. Microbiology 143(Pt 1):93–99
Gibson DT, Subramanian V (1984) Microbial degradation of aromatic hydrocarbons. In: Gibson DT (ed) Microbial degradation of organic compounds. Marcel Dekker, New York, pp 181–252
Goldman R, Enewold L, Pellizzari E, Beach JB, Bowman ED, Krishnan SS, Shields PG (2001) Smoking increases carcinogenic polycyclic aromatic hydrocarbons in human lung tissue. Cancer Res 61:6367–6371
Hansen JB, Olsen RH (1978) Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol 135:227–238
Ka JO, Tiedje JM (1994) Integration and excision of a 2,4-dichlorophenoxyacetic acid-degradative plasmid in Alcaligenes paradoxus and evidence of its natural intergeneric transfer. J Bacteriol 176:5284–5289
Kado CI, Liu ST (1981) Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145:1365–1373
Li W, Shi J, Wang X, Han Y, Tong W, Ma L, Liu B, Cai B (2004) Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 336:231–240
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Mahajan MC, Phale PS, Vaidyanathan CS (1994) Evidence for the involvement of multiple pathways in the biodegradation of 1- and 2-methylnaphthalene by Pseudomonas putida CSV86. Arch Microbiol 161:425–433
van der Meer JR, Sentchilo V (2003) Genomic islands and the evolution of catabolic pathways in bacteria. Curr Opin Biotechnol 14:248–254
van der Meer JR, Ravatn R, Sentchilo V (2001) The clc element of Pseudomonas sp. strain B13 and other mobile degradative elements employing phage-like integrases. Arch Microbiol 175:79–85
Mesas JM, Rodriguez MC, Alegre MT (2004) Plasmid curing of Oenococcus oeni. Plasmid 51:37–40
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Mukhopadhyay M, Mandal NC (1983) A simple procedure for large-scale preparation of pure plasmid DNA free from chromosomal DNA from bacteria. Anal Biochem 133:265–270
Nojiri H, Shintani M, Omori T (2004) Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl Microbiol Biotechnol 64:154–174
Osborn AM, Boltner D (2002) When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48:202–212
Phale PS, Mahajan MC, Vaidyanathan CS (1995) A pathway for biodegradation of 1-naphthoic acid by Pseudomonas maltophilia CSV89. Arch Microbiol 163:42–47
Ravatn R, Studer S, Springael D, Zehnder AJ, van der Meer JR (1998a) Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. Strain B13. J Bacteriol 180:4360–4369
Ravatn R, Zehnder AJ, van der Meer JR (1998b) Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl Environ Microbiol 64:2126–2132
Rheinwald JG, Chakrabarty AM, Gunsalus IC (1973) A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc Natl Acad Sci U S A 70:885–889
Rosas SB, Calzolari A, La Torre JL, Ghittoni NE, Vasquez C (1983) Involvement of a plasmid in Escherichia coli envelope alterations. J Bacteriol 155:402–406
Samanta SK, Rani M, Jain RK (1998) Segregational and structural instability of recombinant plasmid carrying genes for naphthalene degrading pathway. Lett Appl Microbiol 26:265–269
Sambrook JE, Fritsch F, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Springael D, Top EM (2004) Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol 12:53–58
Top EM, Springael D (2003) The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol 14:262–269
Trinder P (1954) Rapid determination of salicylate in biological fluids. Biochem J 57:301–303
Tsuda M, Tan HM, Nishi A, Furukawa K (1999) Mobile catabolic genes in bacteria. J Biosci Bioeng 87:401–410
Weisshaar MP, Franklin FC, Reineke W (1987) Molecular cloning and expression of the 3-chlorobenzoate-degrading genes from Pseudomonas sp. strain B13. J Bacteriol 169:394–402
Wheatcroft R, Williams PA (1981) Rapid methods for the study of both stable and unstable plasmids in Pseudomonas. J Gen Microbiol 124:433–437
Williams PA, Jones RM, Zylstra GJ (2004) Genomics of catabolic plasmids. In: Ramos JL (ed) Pseudomonas: genomics, life style and molecular architecture. Kluwer Academic/Plenum Publisher, New York, pp 165–195
Yen KM, Serdar CM (1988) Genetics of naphthalene catabolism in pseudomonads. Crit Rev Microbiol 15:247–268
Acknowledgements
Thanks to Mr. Balasubramaniyam and Dr. Pradeep Kumar, Naval Materials Research Laboratory, Mumbai for PFGE; Dr. Shouche Y, National Centre Cell Science, India for 16S rRNA analysis. Thanks to Dr. Apte S.K., Bhabha Atomic Research Centre Mumbai; Dr. Balaji P.V., Indian Institute of Technology-Bombay for helpful suggestions. Sophisticated Analytical Instrumentation Facility, IIT-B for GCMS analysis. Aditya Basu would like to thank University Grants Commission for Senior Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Basu, A., Phale, P.S. Conjugative transfer of preferential utilization of aromatic compounds from Pseudomonas putida CSV86. Biodegradation 19, 83–92 (2008). https://doi.org/10.1007/s10532-007-9117-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-007-9117-7